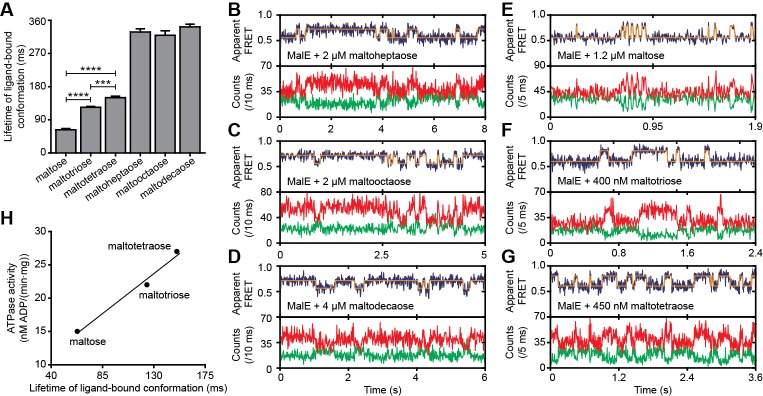
Figure 6. Lifetime of MalE ligand-bound conformations and relation to activity.
(A) Mean lifetime of the ligand-bound conformations of MalE, obtained from all single-molecule fluorescence trajectories in the presence of different maltodextrins as indicated. Data corresponds to mean ± s.e.m. Data in Figure 6—figure supplement 2. Statistical significance was determined by two-tailed unpaired t-tests (***p < 0.005 and ****p < 0.0001). (B, C, D, E, F and G) Representative fluorescence trajectories of MalE(T36C/S352C) in the presence of different substrates as indicated. In all fluorescence trajectories presented: top panel shows calculated apparent FRET efficiency (blue) from the donor (green) and acceptor (red) photon counts as shown in the bottom panels. Most probable state-trajectory of the Hidden Markov Model (HMM) is shown (orange). (H) Published ATPase activity (Hall et al., 1997a) linked to the lifetime of the closed MalE conformation induced by transport of different cognate substrates as indicated. Points are the data and the solid line a simple linear regression fit.
Figure 6—figure supplement 1. Surface-based smFRET histogram of MalE.
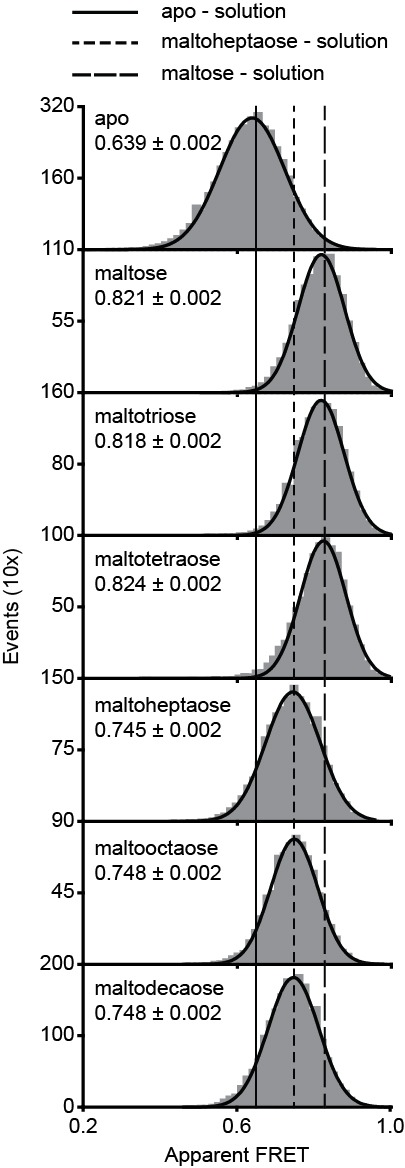
Figure 6—figure supplement 2. Lifetime distribution of the ligand-bound conformations of MalE.
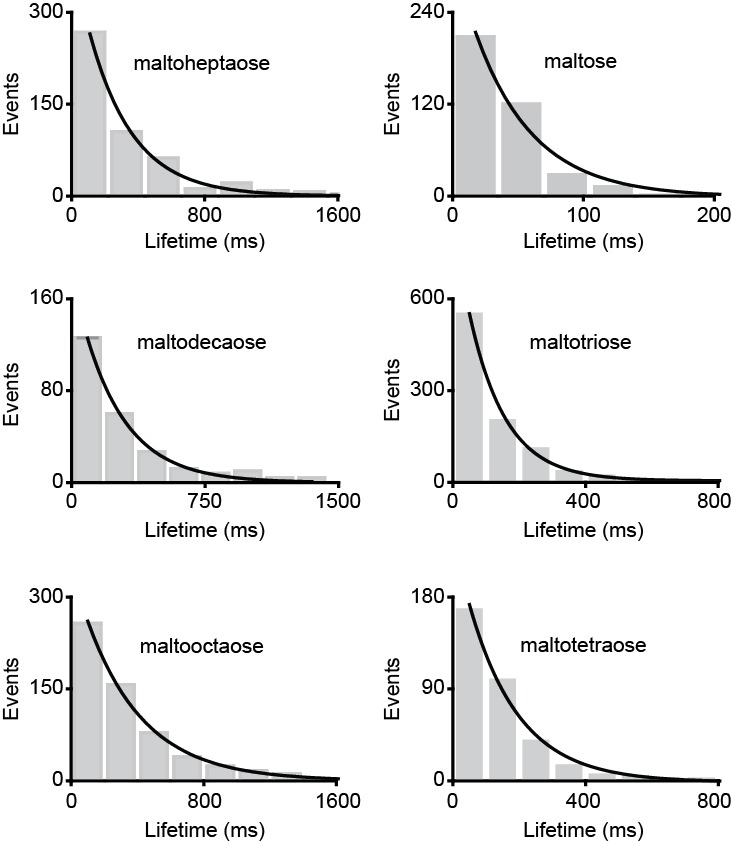
Figure 6—figure supplement 3. Conformational changes and dynamics of MalE(A96W/I329W).