Figure 1.
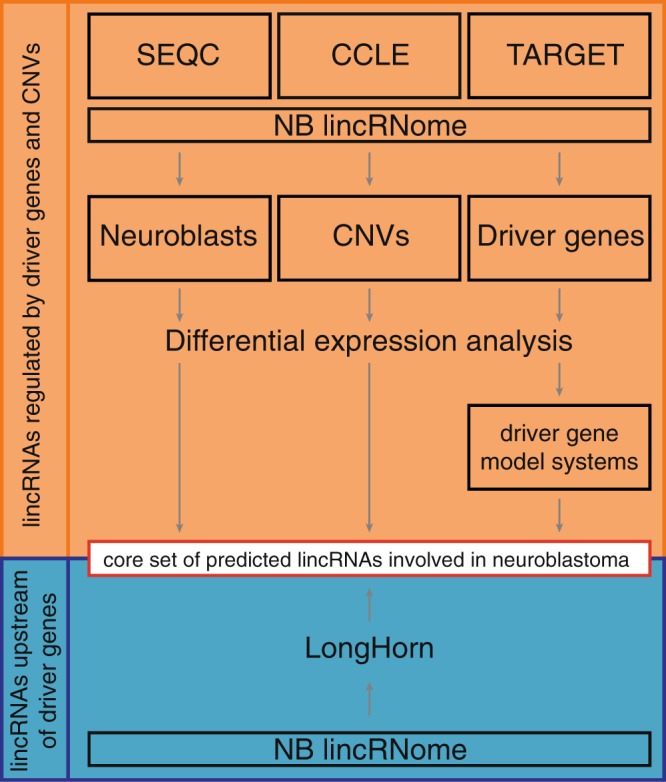
Included data sets and analyses in the study. The SEQC, CCLE and TARGET data sets were used to determine the NB lincRNome. The abundance of the robustly expressed lincRNAs in the lincRNome was used to compare expression between neuroblast and NB samples, primary tumor samples containing CNVs and samples without gains or deletions and NB tumors with and without mutations, amplifications or associations with NB driver genes. The regulation of lincRNAs correlated with NB driver genes was assessed in cellular perturbation models. To determine the involvement of lincRNAs in modulating the effect or regulation of these driver genes, we made use of a state-of-the-art algorithm called LongHorn. Combined, these analyses allow us to arrive at a core set of predicted NB associated lincRNAs.
