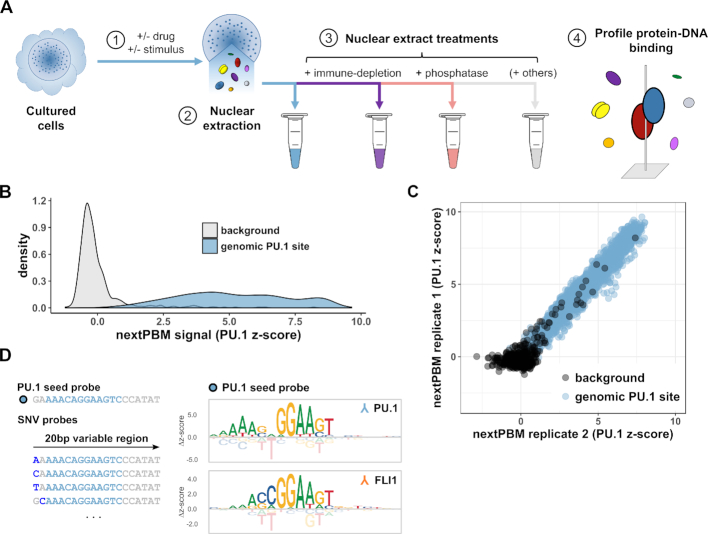
Figure 1.
Nuclear extract protein-binding microarrays (nextPBMs). (A) Workflow schematic for the nextPBM protocol. (1) Cultured cells can be stimulated or treated with a drug prior to nuclear extraction. (2) Total soluble protein content is harvested from cell nuclei using an optimized protocol (see Materials and Methods). (3) Nuclear extract can be treated in parallel enzymatically (i.e. by phosphatase treatment) and components of interest can be depleted (i.e. by immune-depletion using a targeted antibody) depending on goals of the experiments. 4) DNA binding affinity of one or more transcription factors of interest are profiled in parallel directly from nuclear extract. (B) Density of PU.1 nextPBM z-scores obtained at random background probes (n = 500) and at genomic PU.1 binding sites (n = 2615). (C) Scatterplot of PU.1 binding z-scores obtained by DNA probes corresponding to random background (black) and genomic PU.1 sites (blue) in different biological replicates. (D) Left: Schematic representation of the single nucleotide variant (SNV) probes corresponding to an example PU.1 seed probe. Genomic sequence corresponding to the PU.1 motif is highlighted in sky blue within a larger 20bp sequence. SNVs within a given SNV probe are shown in dark blue. Right: Sequence logos obtained for the same genomic PU.1 seed probe using a PU.1 antibody (top) and an FLI1 antibody (bottom). Δz-scores are computed relative to the median score obtained within a given column.