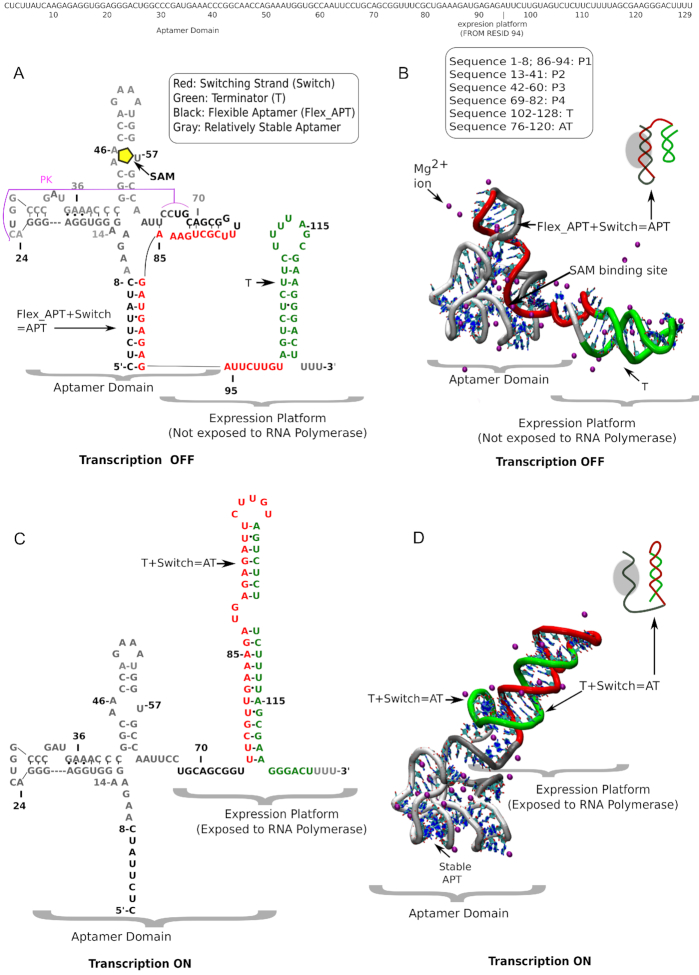
Figure 1.
Secondary and tertiary structure of full-length SAM-I riboswitch (with sequence) in SAM-bound transcription OFF state and SAM-free transcription ON state. (A) Sequence-aligned secondary structure and (B) tertiary structure of the transcription OFF state of SAM-I riboswitch in the presence of metabolite, SAM (yellow pentagon) surrounded by explicit magnesium ions (purple). Different secondary structural segments are defined sequence-wise. Note the partially overlapped aptamer and EP (EP) domains. (C) Sequence-aligned secondary structure and (D) tertiary structures of the transcription ON state surrounded by explicit magnesium (purple) ions. Four characteristic segments, important for switching, are designated with distinct colors: Red: switching strand; green: terminator helix in the EP domain; black: flexible aptamer; gray: more stable aptamer. In the transcription OFF state the flexible aptamer owns the red switching strand. In the transcription ON state green terminator sequesters the red switching strand.