Figure 3.
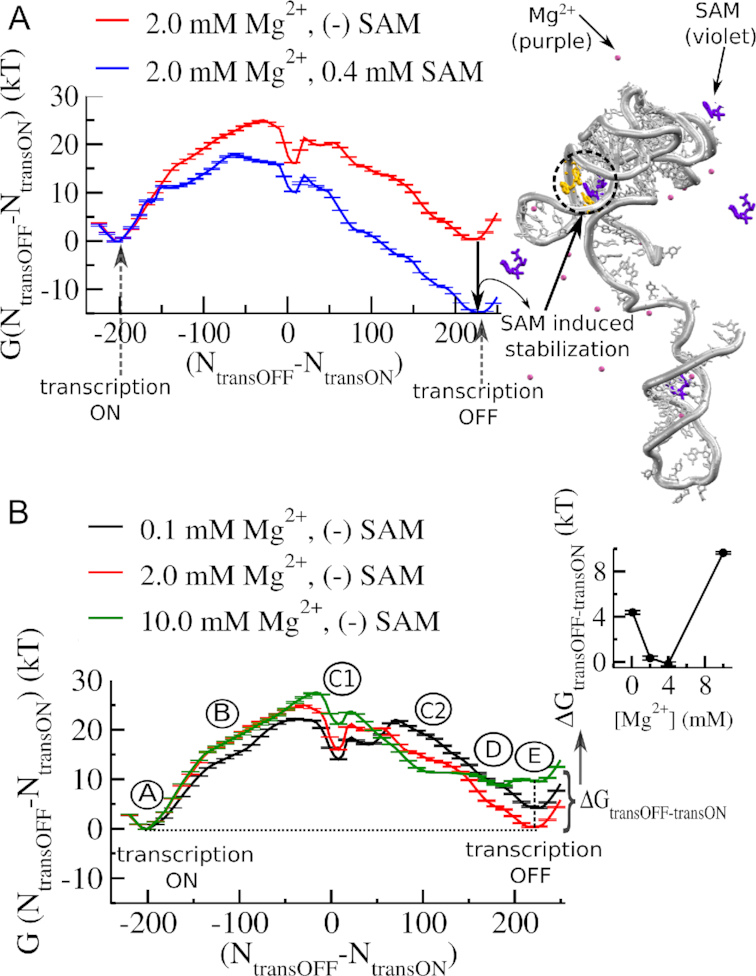
Mg2+ dependence of transcription ON-OFF conformational transition of SAM-I riboswitch. (A) Free energy landscape of transcription ON–OFF transition in the absence and in the presence of metabolite, SAM (0.4 mM). The reaction coordinate is the difference in the number of native contacts that distinguish the transcription ON state (number of contacts between the aptamer and the switching strand (NtransON)) from the transcription OFF state (number of contacts between the terminator and the switching strand (NtransOFF)). Note, SAM-induced stabilization of the transcription OFF state relative to the transcription ON state in the free energy landscape. (B) Mg2+ concentration dependence of transcription ON-OFF transition of SAM-I riboswitch in the absence of SAM. Average free energy profiles at varied [Mg2+] show different stability difference between the transcription ON state and the transcription OFF state. The stability difference between the transcription ON and the transcription OFF states (ΔGtransOFF-transON) quantified as a function of [Mg2+] has a non-monotonic [Mg2+] dependence, as shown in the inset. The aptamer reaches its maximum stability at around 4.0 mM Mg2+ range. Beyond 4.0 mM it starts to destabilize. Along with the stable transcription ON and OFF states (A and E, respectively), the transition involves different intermediates (B, C1, C2, D), which also have different Mg2+ sensitivity.
