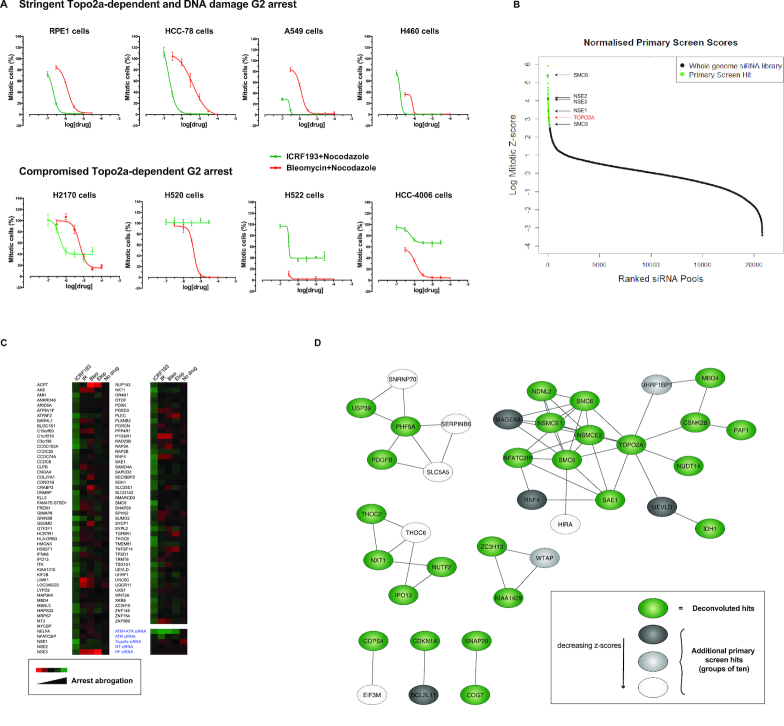
Figure 1.
A genome-wide RNAi screen shows that the Topo2a-dependent G2 arrest is mechanistically distinct from the DNA damage checkpoint and requires the SMC5/6 complex. (A) Graphs showing the responses of the indicated asynchronous cell lines in response to increasing concentrations of ICRF193 or Bleomycin for 24 h. The bypass of arrest population was blocked in mitosis by inclusion of 1 μM Nocodazole and identified by staining for MPM2, yielding a mitotic index (MI) score through automated IF analysis. Data are represented as mean ± S.D. of a representative experiment with eight technical replicates, where the percentage of mitotic cells is normalized to the Nocodazole only treated control. n = 2–3. (B) Distribution of the normalized MI of the genome-wide siRNA library. This data is also part of Supplementary Figure S1E. (C) Heat-map of the arrest abrogation effects of the ICRF193-selected hits in an extended counter-screen with multiple distinct chromatin stresses (IR, ionising radiation; Bleo, bleomycin; Etop, etoposide). (D) The predicted protein-protein interactions for the 48 deconvoluted hits were compiled from the STRING database (green). To this we added additional hits from the primary screen, which were sequentially added in groups of ten based on their ranked z-scores (grey > light grey > white). An additional 30 genes were added before reaching the enrichment score threshold for background noise, 1 × 10−8.