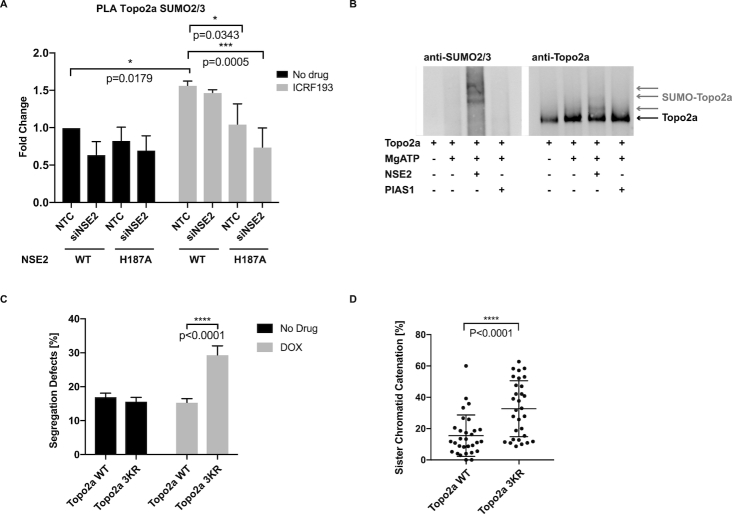
Figure 5.
NSE2-mediated SUMOylation of Topo2a is essential for Topo2a-dependent sister chromatid disjunction. (A) MATLAB-based quantification of PLA experiments using antibodies against Topo2a and SUMO2/3. RPE1 cells stably expressing NSE2wt-GFP or the ligase dead mutant NSE2H187A-GFP were transfected with the indicated siRNAs and treated with 3 μM ICRF193 for 18 h. Data are represented as mean ± S.E.M. (B) WB of in vitro SUMOylation assays using recombinant human Topo2a, SAE1, UBC9, SUMO2 and SUMO3 incubated for 30 min at 37°C with NSE2-GST or PIAS1-GST as indicated. See also Supplementary Figure S6C and D. (C) U2OS FlpIn cells stably expressing inducible Topo2awt-GFP or Topo2a3KR-GFP induced for 48 h with Doxycycline as indicated, fixed and stained with DAPI. Quantification of segregation defects as defined by chromatin bridges or lagging chromosomes. Data are represented as mean ± S.D. n = 3 each with 50 cells. (D) U2OS FlpIn cells as in (C) induced for 24 h with Doxycycline and transfected with siSGO1. The graph shows quantification of SCI in at least 28 cells. Data are represented as mean ± S.D. of a representative experiment. n = 3.