Figure 6.
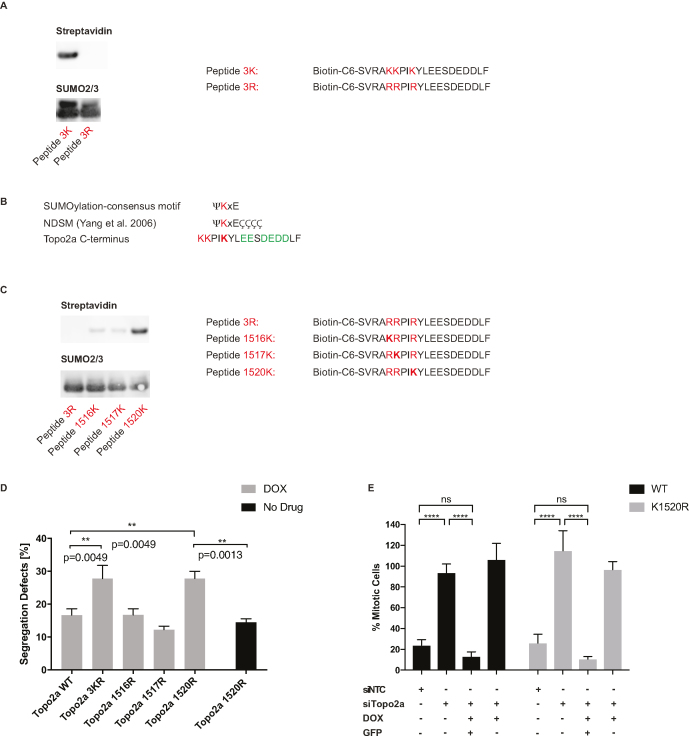
K1520 is a novel NSE2 SUMOylation site on Topo2a. (A) WB of in vitro SUMOylation assays using purified peptides as indicated and SAE1, UBC9, SUMO2 and SUMO3 incubated for 45 min at 37°C with NSE2-GST. The two bands on the SUMO2/3 blot correspond to SUMO2 and SUMO3, which have different molecular weights. Only SUMOylated peptides are large enough to be retained on the SDS-PAGE gel, for transfer and detection by streptavidin-HRP. (B) Comparison of the minimal and the extended SUMO-consensus (NDSM) site with the C-terminus of Topo2a, where ψ represents an amino acid with an alipathic side chain and Ç represents a negatively charged amino acid. (C) Peptide SUMOylation in vitro as in (A). (D) U2OS FlpIn cells stably expressing Doxycycline-inducible GFP-tagged Topo2a, Topo2aK1516R, Topo2aK1517R, Topo2aK1520R or Topo2a3KR induced for 48 h with Doxycycline as indicated, fixed and stained with DAPI. Quantification of segregation defects as defined by chromatin bridges and lagging chromosomes. Data are represented as mean ± S.D. n = 3 each with 30 cells. (E) U2OS FlpIn cells stably expressing doxycycline-inducible GFP-tagged Topo2a WT or Topo2aK1520R were induced for 48 h with Doxycycline and transfected with siRNAs as indicated. Cells were synchronized by a double thymidine block and 5 h after release were treated for 24 h with 3 μM ICRF193 and 1 μM Nocodazole. After fixation cells were stained for GFP, MPM2 and PI. Data are normalized to 1 μM Nocodazole alone and doxycycline induced cells were gated for a GFP positive and negative population, where there were over 1000 GFP positive cells, per condition in each experiment. Data are represented as mean ± S.D. n = 3.