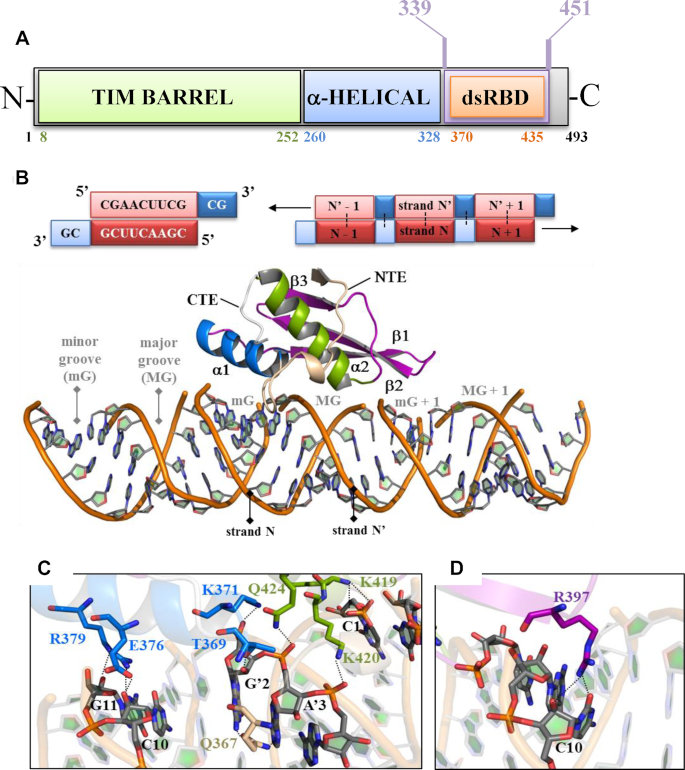
Figure 1.
Mapping dsRBD residues involved in dsRNA recognition by X-ray crystallography. (A) Domain modularity of hDus2. In green is the TIM Barrel domain (TBD), which carries FMN, α-helical domain (HD) is in blue and the extended dsRBD is in purple. Within the latter, there is the canonical dsRBD in orange. (B) On top is shown the RNA palindromic sequence (5′CGAACUUCGCG3′) containing two overhang nucleotides used to generate a dsRNA by self-hybridization. The schematic representation of RNA palindromic sequence and two overhang nucleosides are shown as red and blue boxes, respectively. The 3′–5′ RNA and its 5′–3′ complementary strand are denoted as N and N′ respectively. Dots show the hybridization interface between two complementary sequences while the arrows indicate the 1D direction of macromolecular self-assembly of two complementary strands into a dsRNA helix-A structure. On the bottom is shown the X-ray structure of hDus2 dsRBD in complex with dsRNA. The protein is represented in cartoons wherein helix α1, α2 and β sheet of dsRBD are colored in blue, green and purple, respectively. The N-terminal (NTE) and C-terminal extensions (CTE) are colored in wheat and grey, respectively. dsRNA phosphodiester backbone is represented as an orange ribbon while the ribose and nucleobases are shown as sticks (gray). (C) View on the interactions between (i) residues of helix α1 (blue sticks) and NTE (wheat sticks) and nucleoside from the minor groove (mG) and helix α2 (green sticks) with nucleoside from the major groove (MG) of the dsRNA. The nitrogen, oxygen and phosphate atoms in protein or RNA are in blue, red and orange, respectively. (D) View on the interaction involving R397 (purple sticks) in the C-terminal end of the β1–β2 loop and nucleosides in MG + 1.