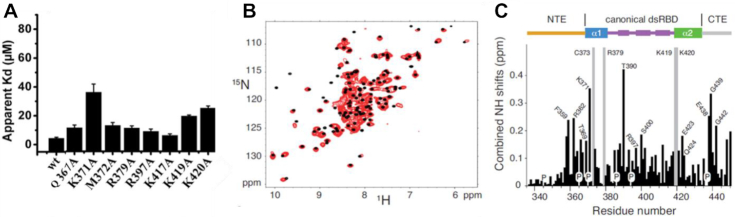
Figure 2.
Characterization of dsRBD-tRNA complex by NMR and mutagenesis. (A) Histogram showing the apparent dissociation constant (Kd) in μM of dsRBD and its mutants for human tRNALys3. (B) shows the (1H,15N)-HSQC experiments of 15N labeled dsRBD free in solution in black or with 1 equivalent of human tRNALys3 in red. (C) Chemical shift perturbation of dsRBD calculated as [(Δδ15N)2/7+(Δδ1H)2]1/2 between its free and tRNA-bound forms. Unobservable residues after titration are shown in grey. Missing values are from proline residues (P) or residues with missing assignment in the free form of dsRBD. The secondary structure of dsRBD is placed above the histogram.