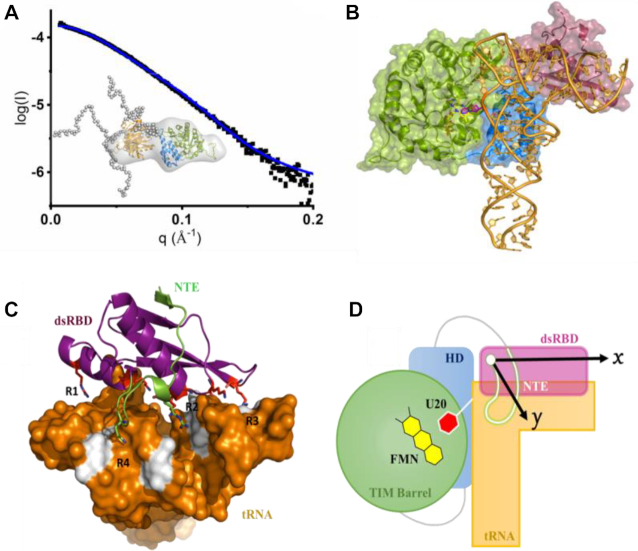
Figure 5.
Molecular model of dsRBD/tRNA complex. (A) SAXS characterization of full length hDus2. Superposition of the experimental (black) and theoretical scattering curves (blue) of hDus2 model; insert: hDus2 model ensemble generated by eom from TBD+HD and dsRBD known X-ray structures superposed on the ab initio shape obtained from dammif. Color code is identical to Figure 1A. In dots are represented the constructed chains by eom. The minimal ensemble best fitting the curve is comprised of 3 components with both DusD and dsRBD having a fixed orientation while flexibility is observed for the C-terminal extremity. (B) Model of full length hDus2/tRNA complex. This model was generated by structural alignment of full length hDus2 with Thermus thermophilus orthologue DusA in complex with tRNA (PDB: 3B0V) (33). TIM Barrel domain, HD and dsRBD are in green, blue and violet respectively. (C) Postulated molecular model for dsRBD/tRNA that is inferred from (C) and supported by compelling biochemical and structural data gathered in this study. tRNA is shown as surface in orange while the canonical dsRBD and NTE are represented as violet and green cartoons, respectively. Colored in white are the nucleotides that are the most impacted by dsRBD binding determined from NMR chemical shift mapping. tRNA recognition by dsRBD is achieved via four regions: R1 to R3 involves the canonical dsRBD while R4 involves only the NTE. (D) Proposed schematic model of full length hDus2-tRNA complex showing the catalytic TIM Barrel domain wherein FMN prosthetic groups (yellow) and the flipped U20 target (red) lye in its center, as a green cycle, HD in blue, dsRBD in violet, NTE in green and tRNA in orange. The arrows show the 2D-surface screened by dsRBD (dsRBD along the x-axis and NTE along the y-axis) acting as a bi-directional interacting adaptor.