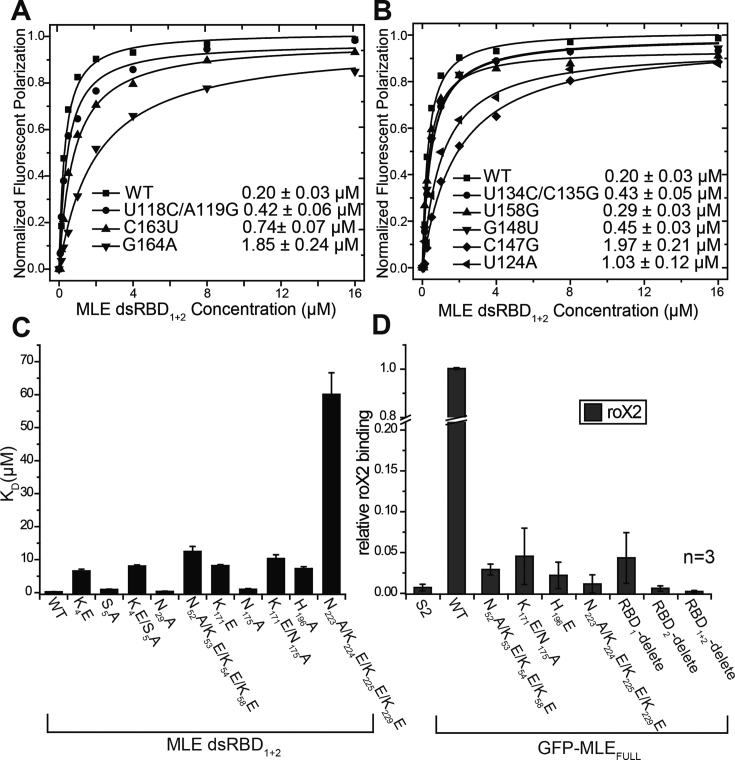
Figure 4.
Effects of structure-based mutations. (A and B) The RNA-binding affinities of MLE dsRBD1+2 for R2H1-wild-type and indicated mutants, determined by FP experiments. Kd values and the corresponding standard errors were determined as described in the ‘Materials and Methods’ section. (C) The histogram of the Kd values shows the RNA-binding affinities of R2H1 for MLE RBD1+2-wild-type and indicated mutants, as determined by FP experiments. (D) RIP assays performed with GFP-tagged MLEFL-wild-type and indicated mutants. The abundance of roX2 RNA was quantified by qRT-PCR using specific primers for roX2. Relative roX2 enrichment (IP/input) in mutants was normalized to wild-type, while the relative roX2 enrichment (IP/input) in wild-type was set to 1.0. Data are presented as the means ± SD. n represents the number of experiments.