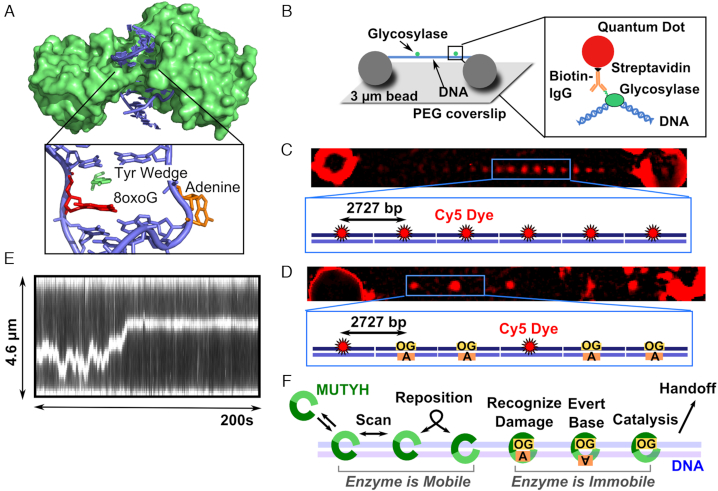
Figure 1.
MutY structure, single-molecule assay, and model for MUTYH diffusion. A) Crystal structure of B. stearothermophilus MutY (green) (PDB 1RRQ) crosslinked to an 8-oxoG:A site showing the enzyme encircling the DNA (blue). The wedge residue (green) intercalates 5′ to the damage (red), and the adenine (orange) is everted from the base stack into the active pocket. B) DNA molecules (blue) extended by hydrodynamic flow and non-specifically attached between immobile 3 μm polylysine-coated silica beads in a flow cell to resemble tightropes for observation of glycosylase (green) scanning. Qdot-glycosylase complex constructed by targeting the hexahistidine tag on the C-terminus of the glycosylase with a biotinylated antihistidine IgG linked to the streptavidin-coated Qdot. (C) DNA tightropes consist of concatemerized plasmids containing regularly spaced Cy5 fiducial markers 2727 base pairs apart. (D) Tightrope substrates for single molecule experiments consist of a combination of Cy5 marked plasmids and plasmids that contain a single damage site (8-oxoG:A or 8-oxoG:C). The substrates concatemerize with randomly distributed Cy5 markers, and the position of the damage site can be determined using the plasmid repeat distance (905 nm). (E) Example kymograph of MUTYH scanning along a concatemer tightrope that contains 8-oxoG damage sites. (F) Model for MUTYH scanning along tightrope DNA.