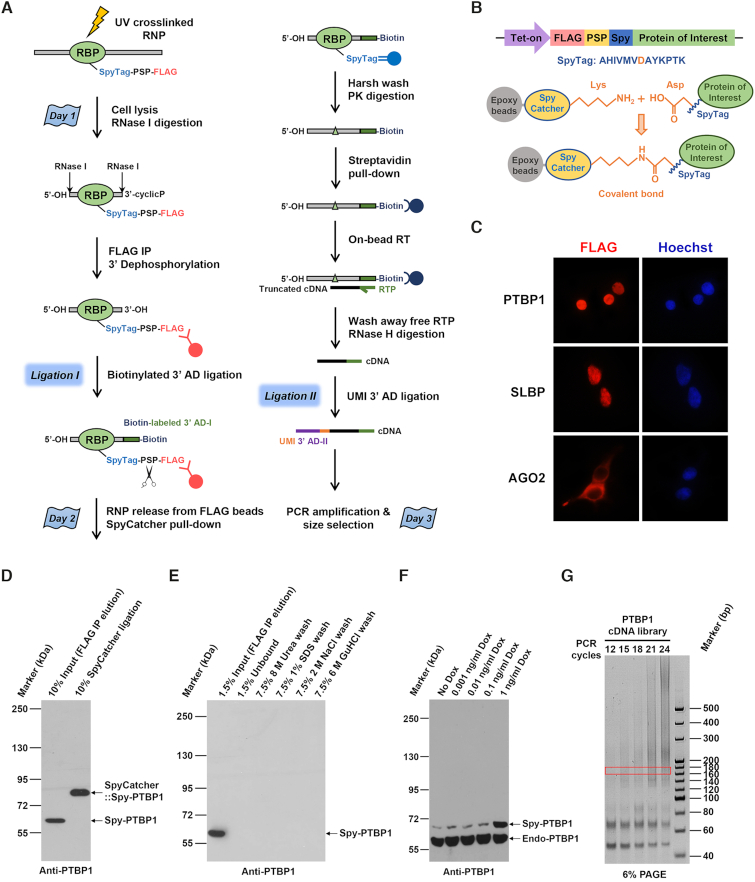
Figure 1.
A highly efficient covalent linkage-based SpyCLIP platform for the characterization of protein–RNA interactions. (A) Schematic representation of the SpyCLIP procedure. (B) Schematic representation of the Spy-tagged RBP structure and mechanism of the SpyTag-SpyCatcher covalent interaction. (C) Cellular localization of Spy-tagged RBPs ectopically expressed in Lenti-X 293T cells. (D) The modified SpyCatcher protein efficiently formed a stable complex with Spy-tagged PTBP1 in SpyCLIP lysis buffer. (E) The covalent SpyTag-SpyCatcher pull-down system can withstand harsh washing conditions, including buffers containing 8 M urea, 1% SDS, 2 M NaCl or 6 M guanidine hydrochloride. (F) Comparison of expression levels of Dox-induced Spy-tagged PTBP1 with its endogenous counterpart in Lenti-X 293T cells, and 1 ng/ml Dox was used for all RBPs used in this study. The PTBP1 band in the No Dox lane is due to leak expression of the tet-on system. (G) Amplification of PTBP1 SpyCLIP libraries using different PCR cycles. Twenty PCR cycles can provide a sufficient amount of cDNA for deep sequencing in a relatively small volume and did not increase the percentage of PCR duplicates; hence, 20 PCR cycles were chosen for library preparations. The region marked by the rectangle (160–180 bp) was recovered for deep sequencing.