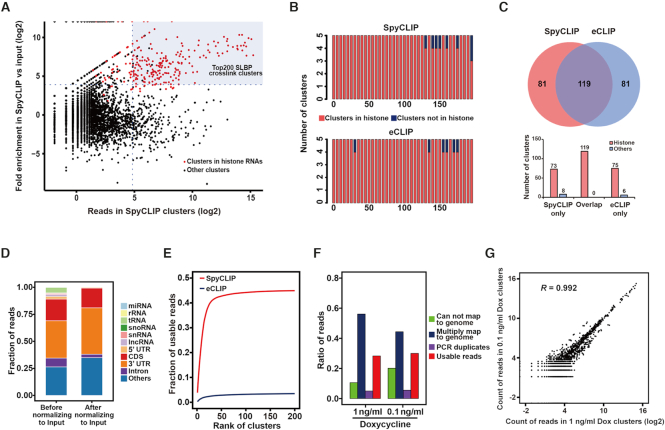
Figure 3.
Improved SpyCLIP specificity and sensitivity after background deduction by a universal input library. (A) Fold enrichment for SLBP SpyCLIP clusters relative to the universal input library. The dots in red indicate the clusters overlapping with histone mRNA. The fold enrichment is defined as 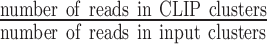 , and clusters with a fold enrichment > 15 are defined as SLBP-specific ones. (B) Histograms show the number of clusters located within histone mRNAs in the top 200 SLBP-specific clusters identified by SpyCLIP or eCLIP. The clusters were ranked and binned by their expression levels. (C) Overlap of SLBP-specific clusters identified by SpyCLIP and eCLIP. The number of clusters within histone mRNA or nonhistone RNAs in each section are shown in the lower panel. (D) Genomic distribution of SLBP SpyCLIP clusters before and after input deduction. Most of the noise signals removed by input normalization originated from abundant endogenous rRNAs, tRNAs, snRNAs and snoRNAs. (E) The cumulative fraction of usable reads in the top 200 SLBP-specific clusters identified by SpyCLIP or eCLIP. (F) Composition of sequencing reads from SLBP SpyCLIP libraries at different Dox induction concentrations. (G) Reproducibility of SLBP SpyCLIP reads within the identified clusters from libraries at different Dox induction concentrations.
, and clusters with a fold enrichment > 15 are defined as SLBP-specific ones. (B) Histograms show the number of clusters located within histone mRNAs in the top 200 SLBP-specific clusters identified by SpyCLIP or eCLIP. The clusters were ranked and binned by their expression levels. (C) Overlap of SLBP-specific clusters identified by SpyCLIP and eCLIP. The number of clusters within histone mRNA or nonhistone RNAs in each section are shown in the lower panel. (D) Genomic distribution of SLBP SpyCLIP clusters before and after input deduction. Most of the noise signals removed by input normalization originated from abundant endogenous rRNAs, tRNAs, snRNAs and snoRNAs. (E) The cumulative fraction of usable reads in the top 200 SLBP-specific clusters identified by SpyCLIP or eCLIP. (F) Composition of sequencing reads from SLBP SpyCLIP libraries at different Dox induction concentrations. (G) Reproducibility of SLBP SpyCLIP reads within the identified clusters from libraries at different Dox induction concentrations.