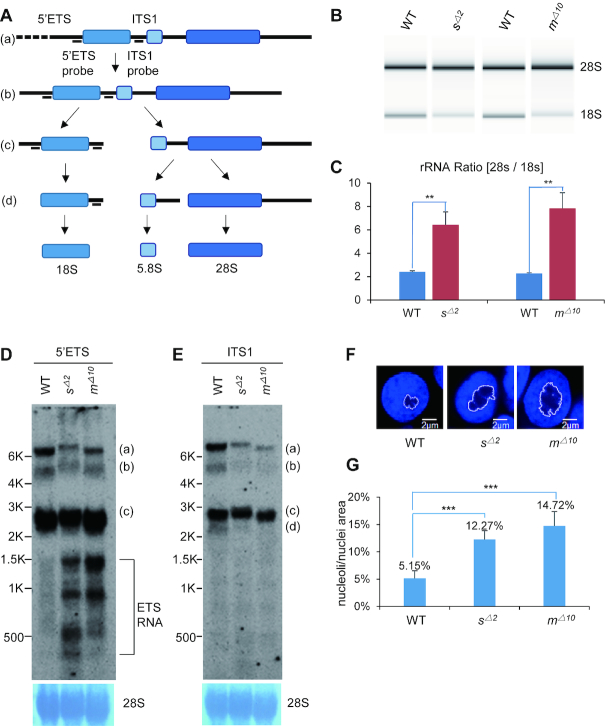
Figure 2.
Depletion of Sas10 and Mpp10 leads to pre-rRNA processing defects. (A) Diagram showing the stepwise processing of the pre-rRNA transcript (a) to the intermediates (b and c) to the mature 18S rRNA. 5′ETS, 5′-external transcribed sequence; ITS1, internal transcribed sequence. (B and C) Detection of 28S and 18S rRNA on an Agilent 2100 Bioanalyzer (B) for analysing the ratio of 28S rRNA versus 18S rRNA (C) in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) embryos.**, P< 0.01 (D and E) Northern blot analysis of rRNA precursors using the 5′ETS (D) and ITS1 (E) probes in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) embryos. 28S: loading control. (F and G) DAPI was used to stain the nuclei. The area of nucleolus (regions lacking or showing faint DAPI signal) and of nucleus in an individual cell was measured (F), respectively, in 5dpf-old WT, sas10Δ2 (sΔ2) and mpp10Δ10 (mΔ10) liver. The average area ratios of nucleolus/nucleus from at least 90 hepatocytes from three embryos for each genotype were used in statistic analysis (G); bar, 2 μm. ***, P < 0.001.