Figure 7.
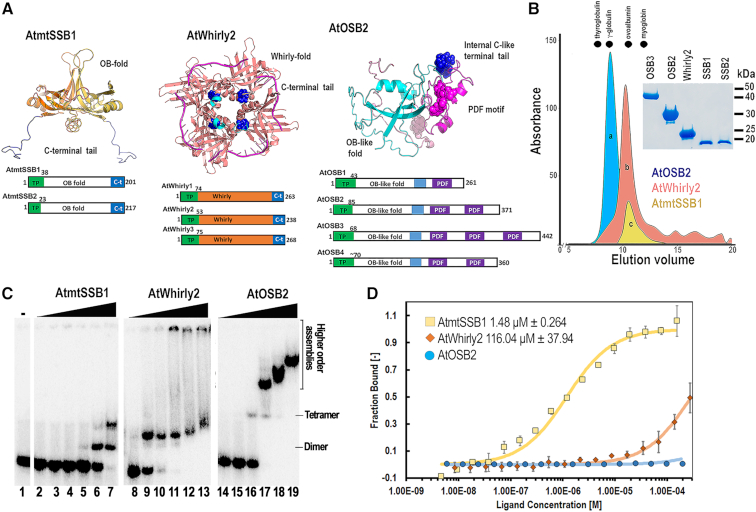
Arabidopsis harbors canonical and unique SSBs. (A) Structural organization of AtmtSSBs, AtWhirlies and AtOSBs. AtmtSSB1 and AtmtSSB2 are predicted to assemble as proteins with an oligonucleotide/oligosaccharide-binding (OB)-fold and a C-terminal disorder region, AtWhirlies (1–3) consists of a Whirly domain composed of two four-stranded β-sheets and two α–helices plus a disordered C-terminal extension (55,56), AtOSBs consist of an OB-like fold followed by one to three PDF motifs (54). The structural models of AtmtSSBs and AtOSBSs highlight the structural organization of the OB-fold domains and PDF motifs respectively. Structural modeling shows AtmtSSB1 and AtOSB1 as a dimer and monomer for clarity. The crystal structure of AtWhirly2 is shown in combination with the superimposed ssDNA co-crystallized in whirly2 from potato. The C-terminal amino acids of Whirlys are not present in their crystal structures and are proposed to be involved in protein-protein interactions. The AtWhirly2 structure illustrates the tetrameric organization and DNA binding of members this protein family. (B) Gel filtration profiles of purified AtmtSSB1, AtOSB2, and AtWhirly2 (inset) on a Superdex 75 column. Dots denote the elution profiles of molecular weight markers: thyrogloublin, γ globulin, ovoalbumin and myoglobin. The elution profiles of AtSSBs indicate their tetrameric assembly. (C) EMSA assay showing the binding of purified AtmtSSB1, AtOSB2, and AtWhirly2 to ssDNA. Reactions were incubated with increased concentration of the tetrameric SSBs (0, 1.15, 4.125, 14.87,53.5, 192.9, 694.2 nM) and 1 nM of a 5′-32P-labeled 70-mer oligonucleotide. Binding reactions were resolved on an 8% non-denaturing polyacrylamide gel. (D) Langmuir isotherms showing the binding of AtmtSSB1 and AtWhirly2 to AtPolIB exo− by MST.