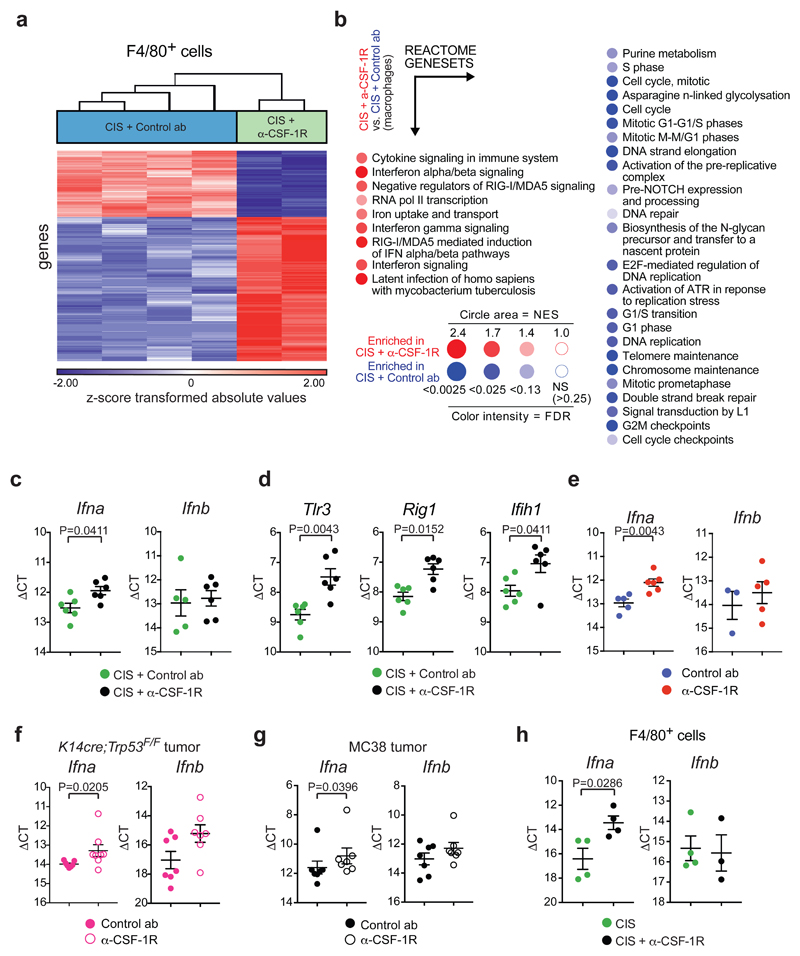
Figure 3. CSF-1R inhibition alters TAM phenotype and induces type I IFN signaling in the tumor microenvironment.
(a) Hierarchical clustering of the top 400 variable genes between CD11b+F4/80+ cells isolated from KEP tumors treated with cisplatin/control ab (n=4 animals) and cisplatin/anti-CSF-1R (n=2 biologically independent samples, pool of 5 mice each). FC: ≥ 1,5; p-value with FDR correction: ≤ 0,05. Statistical analysis was performed using two-way ANOVA. (b) BubbleGUM visualization of GSEA using reactome genesets comparing CD11b+F4/80+ cells from cisplatin/anti-CSF-1R-treated tumors versus macrophages from cisplatin/control ab-treated tumors. Same mice as in a. (Normalized enrichment score (NES) ≥ 1, FDR value: 0.25 ≤). Enrichments score was calculated using a weighted Kolmogorov–Smirnov-like statistic. (c) Transcripts of Ifna (n=6 animals/group) and Ifnb (n=5 animals in cisplatin/control ab, n=6 animals in cisplatin/anti-CSF-1R) and (d) Tlr3, Rig1 and Ifih1 in KEP mammary tumors isolated one day after the second cisplatin injection were determined by qPCR and normalized to β-actin (n=6 animals/group). (e-g) Transcripts of Ifna and Ifnb in KEP mammary tumors (Ifna: n=5 animals in control ab, n=6 animals in anti-CSF-1R; Ifnb: n=3 animals in control ab, n=5 animals in anti-CSF-1R) (e), orthotopically transplanted K14cre;Trp53F/F mammary tumor (Ifna: n=7 animals in control ab, n=8 animals in anti-CSF-1R; Ifnb: n=7 animals/group) (f) and subcutaneously inoculated MC38 tumors (n=7 animals/group) (g) treated with control ab and anti-CSF-1R were determined by qPCR and normalized to β-actin. Mice were analysed at a tumor size of 100mm2 (KP) or after 12 days from the start of the treatment (MC38). (h) Transcripts of Ifna (n=4 animals/group) and Ifnb (n=4 animals in cisplatin, n=3 animals in cisplatin/anti-CSF-1R) in CD11b+F4/80+ cells isolated from end-stage KEP tumors were determined by qPCR and normalized to β-actin. Graphs in c-h show the mean ± SEM in ΔCt values and statistical analysis was performed using two-tailed Mann–Whitney test.