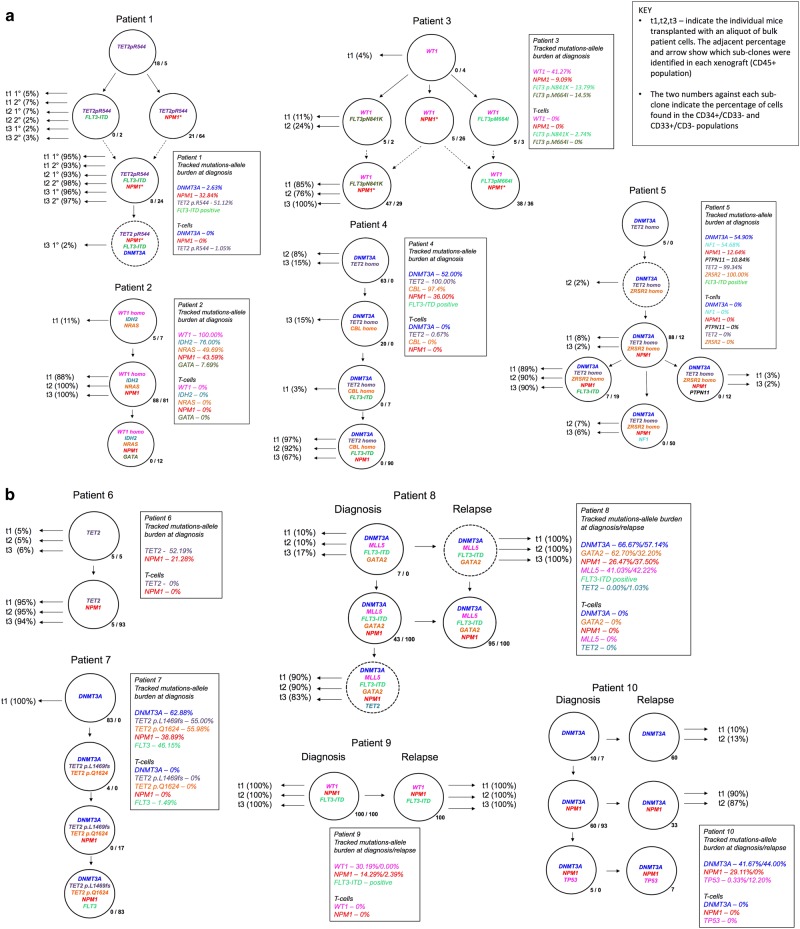
Fig. 1.
Clonal phylogenies, inferred by maximum parsimony, and sub-clone genotypes in 10 patients. Genetically distinct sub-clone percentages (as a fraction of the total population) are indicated next to each clone; e.g., patient 1, most primitive sub-clone, CD34+/CD33− first and CD33+/CD34-/CD3− second percentages indicated as 18%/5%, respectively. This indicates that this sub-clone was found in 18% of the total CD34+/CD33- cells investigated and 5% of the total CD33+/CD34−/CD3−cells investigated (for the relapse samples of patients 9 and 10 only bulk cells without phenotype consideration could be sorted, as the samples available were from fixed cytogenetic preparations; the sub-clone is shown as a single percentage). Those sub-clones that grew in mice are indicated with horizontal black arrows. t1-3 (%). T, transplant. 1-3 individual mice. % fraction of human cells in mouse bone marrow. Sub-clone denoted by dotted circle is below detection limit in diagnostic sample but present in mouse transplant read-out. Dotted arrows lines between sub-clones (case #1 and #3) indicates alternative clonal phylogenies. In case #3, there are 4 possible equally parsimonious phylogenetic trees (details in Supplementary Information Figs. 5 and 6). Further details on each of the individual 10 patients’ clonal analyses are given in Supplementary Information