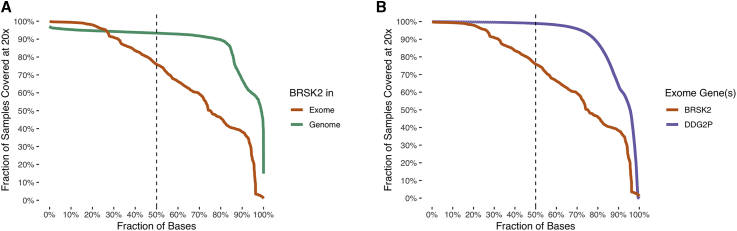
Figure 3.
Comparisons of BRSK2 Sequencing Depth across gnomAD Datasets
Fractions of gnomAD samples that attain a per-base sequencing depth of ≥20× are plotted as a function of the percentage of bases examined, ordered by a decreasing fraction of exonic-base coverage. Only autosomal positions are included. The dashed line shows the fraction of samples covered at the median-depth base.
(A) Using only BRSK2 exonic bases (exons plus 10 bp on either side), coverage is compared in gnomAD exomes (orange; 125,748 individuals) and gnomAD genomes (green; 15,708 individuals).
(B) Using only gnomAD exomes (125,748 individuals), exonic bases (exons plus 10 bp on either side) in BRSK2 (orange) are compared to exonic bases in 1,012 confirmed developmental-delay genes identified by the Developmental Disorders Genotype-Phenotype Database (DDG2P; purple).