Capsule Summary:
In this large, multi-ethnic GWAS of asthma, we identified novel associations with potential functional relevance for asthma susceptibility in older adults of diverse racial backgrounds.
Keywords: asthma, large-scale GWAS, GERA, adult
To the Editor:
Asthma is a genetically heterogeneous disease. While many recent genome-wide association studies (GWAS) have implicated multiple loci associated with asthma risk1–4, the impact of genetic variation on asthma susceptibility in large, ethnically diverse human populations has not been fully ascertained. Identifying the genetic variants associated with asthma in well-powered GWAS is crucial for discerning the genetic basis of asthma. Furthermore, it is also necessary to understand how genetic heterogeneity underlying asthma risk may be influenced by ethnic background. The objective of this study was to conduct a large-scale, trans-ethnic GWAS of asthma in individuals in the Kaiser Permanente Northern California Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort5,6.
Demographic information for GERA (68,623 asthma cases and non-asthmatic controls) is provided in Supplemental Table 1. GWASs were conducted for four racial/ethnic groups within GERA (non-Hispanic white, African-American, Asian, and Hispanic), and replicated using data from two large consortia, EVE and GABRIEL. Selected SNP associations with potential functional consequences were profiled using bioinformatic methods. All study procedures were approved by the Institutional Review Board of the Kaiser Foundation Research Institute and Brigham and Women’s Hospital. A detailed Methods section is provided in this article’s Online Repository at www.jacionline.org.
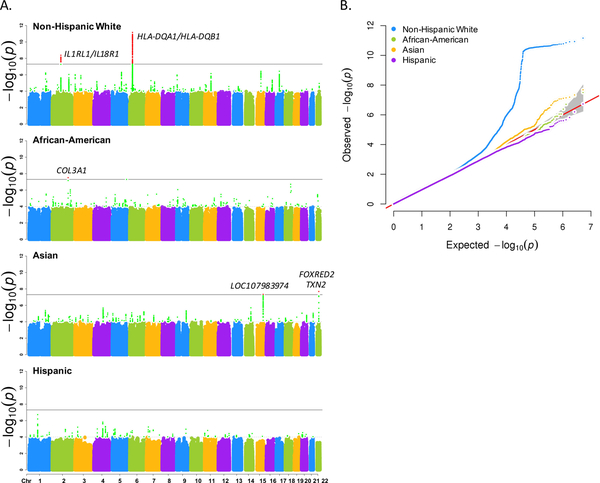
In non-Hispanic whites, >7 million SNPs passed QC measures and were evaluated by GWAS, of which 646 SNPs met genome-wide significance (P<5×10−08) and were annotated to two regions of chromosomes 2 and 6 that include IL1RL1/IL18R1 and HLA-DQA1/HLA-DQB1, respectively; of these, five SNPs were also genotyped in GABRIEL and four were suggestive in GABRIEL (P<1×10−05) (Fig. 1A; Table 1). Eighteen of the genome-wide significant GERA SNPs were also genotyped in EVE; of these, 14 were nominally significant (P<0.05), and the top two SNPs were suggestive (P<1×10−05) (Table 1). Thus, a total of 16 GERA SNPs replicated in either EVE or GABRIEL, of which all were annotated to HLA-DQA1 or IL1RL1/IL18R1 loci (Fig. 1A; Table 1). A survey of 32 published asthma GWASs cited in the NHGRI Catalog revealed 113 suggestive associations (P<1×10−05) (Supplemental Table 2). Of these, 87 SNPs (77%) were also genotyped in the GERA non-Hispanic white dataset, of which 25 replicated at a Bonferroni-corrected P-value threshold of 4.42×10−04. When the GERA SNP P-values were combined with those of the cited NHGRI studies, 44/87 (51%) SNPs in 28 unique loci met the Bonferroni-corrected cutoff; of these, 25 SNPs in 16 loci met the traditional genome-wide significance threshold of P<5×10−08 (Supplemental Table 2).
Figure 1. Discovery GWAS Results for GERA Populations.
A. Manhattan plots for each GWAS are provided by racial/ethnic group, with chromosomal positions along the abscissa while –log(p-value) is shown on the ordinate axis. Horizontal lines indicate significance thresholds for genome-wide (black line) and suggestive genome-wide associations (dashed line), respectively. B. Q-Q plot of results for each GWAS. For all populations, λgc values were 1.0–1.004.
Table 1.
GWAS Results for Non-Hispanic Whites
| SNP | Chr. | Location (bp)* | Gene | Minor Allele* |
Discovery (GERA) | Replication (GABRIEL) | Replication (EVE) | Combined P-Value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | ||||||
| rs17843604 | 6 | 32620283 | HLA-DQA1 | C | 0.90 (0.88−0.93) | 1.10×10−10 | 0.86 (0.82−0.90) | 1.68×10−10 | --- | --- | 1.39×10−18 |
| rs17612633 | 6 | 32615710 | HLA-DQA1 | G | 0.89 (0.87−0.93) | 1.36×10−11 | 0.87 (0.81−0.93) | 4.07×10−05 | --- | --- | 2.59×10−15 |
| rs1420101 | 2 | 102957716 | IL1RL1 | T | 1.10 (1.06−1.13) | 2.65×10−08 | 1.11 (1.06−1.15) | 9.60×10−07 | 1.15 (1.0620131.25) | 2.87×10−05 | 8.25×10−15 |
| rs12998521 | 2 | 102974417 | IL18R1 | T | 1.10 (1.06−1.13) | 1.34×10−08 | 1.11 (1.07−1.16) | 1.39×10−07 | 1.10 (1.0120131.20) | 1.42×10−03 | 3.05×10−14 |
| rs9272346 | 6 | 32604372 | HLA-DQA1 | G | 0.89 (0.97−0.93) | 1.13×10−11 | --- | --- | 0.93 (0.8520131.00) | 4.29×10−04 | 1.50×10−12 |
| rs9272723 | 6 | 32609427 | HLA-DQA1 | T | 0.89 (0.87−0.93) | 2.52×10−11 | --- | --- | 0.92 (0.8520131.00) | 1.58×10−03 | 4.46×10−12 |
| rs13019081 | 2 | 102950822 | IL1RL1 | C | 1.11 (1.07−1.14) | 2.09×10−09 | --- | --- | 1.11 (1.0220131.22) | 3.41×10−04 | 3.28×10−10 |
| rs12712142 | 2 | 102960584 | IL1RL1 | A | 1.01 (1.07−1.14) | 4.65×10−09 | --- | --- | 1.13 (1.0520131.23) | 7.15×10−05 | 5.94×10−10 |
| rs13001325 | 2 | 102939036 | IL1RL1 | T | 1.11 (1.07−1.14) | 3.49×10−09 | --- | --- | 1.11 (1.0220131.22) | 7.63×10−04 | 6.43×10−10 |
| rs12479210 | 2 | 102949161 | IL1RL1 | T | 1.11 (1.07−1.14) | 4.32×10−09 | --- | --- | 1.11 (1.0220131.22) | 3.34×10−04 | 6.98×10−10 |
| rs11123923 | 2 | 102967844 | IL1RL1 | A | 1.10 (1.07−1.14) | 3.89×10−09 | --- | --- | 1.1 (1.0120131.2) | 8.32×10−04 | 7.30×10−10 |
| rs950880 | 2 | 102932562 | IL1RL1 | A | 1.10 (1.07−1.14) | 5.66×10−09 | --- | --- | 1.11 (1.0220131.21) | 1.88×10−04 | 8.45×10−10 |
| rs6543119 | 2 | 102963072 | IL1RL1 | T | 1.10 (1.06−1.13) | 7.15×10−09 | --- | --- | 1.14 (1.0520131.23) | 1.23×10−04 | 1.01×10−9 |
| rs13017455 | 2 | 102964742 | IL1RL1 | T | 1.10 (1.06−1.13) | 7.53×10−09 | --- | --- | 1.14 (1.0420131.23) | 2.50×10−04 | 1.19×10−9 |
| rs2287037 | 2 | 102979028 | IL1RL1 | T | 1.10 (1.06−1.13) | 1.14×10−08 | --- | --- | 1.10 (1.0220131.22) | 6.01×10−03 | 3.22×10−9 |
| rs9269080 | 6 | 32440969 | HLA-DRB9 | G | 0.91 (0.9−0.95) | 2.39×10−08 | --- | --- | 0.94 (0.8820131.02) | 0.04 | 1.06×10−8 |
Table shows the SNP identifiers, gene annotations, GWAS summary statistics and joint P-values for GERA, GABRIEL and EVE.
Chromosomal location and minor allele designations for GERA.
In the three additional racial/ethnic groups in GERA, 6.4–12.7 million SNPs passed QC and were evaluated by GWAS. Three SNPs (rs5756210 downstream of FAD dependent oxidoreductase domain containing 2 (FOXRED2), rs10534662 in thioredoxin 2 (TXN2), and rs8042955 in LOC107983974/RP11–739G5.1) achieved genome wide significance in Asians, and rs13306275 in collagen type III alpha 1 chain (COL3A1) was genome wide significant in African Americans (Fig. 1A; Supplemental Table 3). No genome-wide significant or suggestive associations replicated for these groups in EVE, GABRIEL or the NHGRI database. In addition, none of the associations approaching genome-wide significance overlapped with any genome-wide significant/suggestive SNP association across all four racial/ethnic groups.
We attempted to discern the potential functional relevance of the most highly significant genome-wide-significant SNPs. The top-ranked SNP in non-Hispanic whites was rs9272513, which is present in an intron of HLA-DQA1 and is also an eQTL for both HLA loci and IL1RL1/IL18R1 in the lung and circulation (Supplemental Table 4). In addition, rs12712142 was associated with an 11% increased risk of asthma and is an eQTL for IL1RL1, reducing transcript expression in the lung (Supplemental Fig. 1). Further characterization of rs12712142, which was annotated to the 3’UTR region of IL1RL1, showed that it introduces a new binding site for hsa-miR-452–5p, which is also significantly up-regulated in asthmatic airway epithelia compared to samples from healthy, non-atopic controls (Supplemental Fig. 2; Supplemental Fig. 3). Since variations across multiple genes within affected pathways could directly impact asthma phenotypes, we analyzed 2,143 suggestive (P<1×10−05) associations from non-Hispanic whites in GERA for gene-set and tissue enrichment. Tissue and cell enrichment for relevant gene sets included those related to CD4+, CD8+, B and T-cell growth, morphology and other activities while seven tissue enrichment sets included pathways for ‘T-lymphocytes’ and ‘CD4 Positive T-lymphocytes’ (Supplemental Table 5).
Collectively, these analyses replicated previously reported top asthma GWAS findings as well as increased the significance threshold for many of these associations, in addition to expanding the list of novel SNPs that are strongly associated with asthma risk. Due to differences in GWAS study design, imputation, and available SNP data, the majority of genome-wide significant SNPs from the discovery GWAS were not well-represented in EVE or GABRIEL, limiting the total number of SNPs for replication. However, there was a moderate to high level of LD between the top discovery GWAS SNPs not genotyped in GABRIEL or EVE and the replicated SNPs shown in Table 1. Conditioning on the lead replicated SNP for chr6, rs17843604, reduced the strength of the regional GWAS association of HLADQA1 (within 400 kb) to below genome-wide significant levels (top SNP unadjusted P value = 7.5×10−07) (Supplemental Figure 4). Similarly, conditioning on rs1420101 removed all significant associations within 400 kb of the IL1RL1/IL18R1 region. Finally, while we present novel GWAS findings from racial/ethnic groups in addition to non-Hispanic whites, our ability to replicate associations from these and other populations was limited, due in part to the aforementioned reasons. A recently conducted GWAS of adults and children with and without asthma also revealed novel associations in 6p21.31, 9p21.2, and 10q21.3 in European-Americans and 9q34.11 in African-Americans, but we did not replicate these in our analysis of GERA7. Nevertheless, like Almoguera et al., we did capture suggestive associations for established asthma susceptibility loci in individuals of European ancestry, including ORMDL3/GSDMB, IL33, and others.
The lack of overlap of asthma associations across the four ethnic groups in GERA may have more than one underlying explanation. One possibility is that we did not have sufficient statistical power to detect the same genetic associations across all racial/ethnic subgroups, due to variance in minor allele frequencies and lower sample sizes for the minority participants. However, differences in associations across racial/ethnic groups could also be attributed to unique biological pathways involved in asthma susceptibility in these populations. In Asians, asthma case status was associated with SNPs annotated to two oxidative stress pathway genes, FOXRED2 and TXN2. Neither SNP was previously reported as associated with asthma in any population, and therefore could represent unique genetic risk factors in individuals of Asian ancestry. (Of note, the FOXRED2 variant was not genotyped in African Americans, and therefore could not be tested in our GWAS). In Hispanics, we observed suggestive associations with asthma for five unique genes, including TET2, which is associated with COPD phenotypes; again, none were previously associated with asthma risk. Finally, SNPs in collagen- and cadherin-related genes also showed potential relationships with asthma in African-American subjects. While other GWASs have reported associations between collagen8 and cadherin genes9 and asthma phenotypes, this is the first GWAS to specifically identify COL25A1, COL3A1 and CDH7 as potential candidates.
In conclusion, previously unreported SNPs within the MHC II region of chromosome 6 and the IL1RL1/IL18R1 region of chromosome 2 are associated with asthma susceptibility in adults of non-Hispanic white ancestry, while novel SNPs in FOXRED2 and TXN2 are associated with asthma in Asians, and COL3A1 is associated with asthma in African-Americans. One of the top associations, rs12712142, could confer a novel potential mechanism for asthma susceptibility through introduction of a new miRNA binding site that regulates expression of IL1RL1. This study contributes novel associations with potential functional relevance for asthma susceptibility in older adults of diverse racial backgrounds, and represents one of the largest multi-ethnic GWAS of asthma to date.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19(4):458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S et al. A large-scale, consortium-based genomewide association study of asthma. The New England journal of medicine. 2010;363(13):1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362(1):36–44. [DOI] [PubMed] [Google Scholar]
- 4.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almoguera B, Vazquez L, Mentch F, Connolly J, Pacheco JA, Sundaresan AS et al. Identification of Four Novel Loci in Asthma in European American and African American Populations. Am J Respir Crit Care Med. 2017;195(4):456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan QL, Lasky-Su J, Himes BE, Qiu W, Litonjua AA, Damask A et al. A genome-wide association study of bronchodilator response in asthmatics. Pharmacogenomics J. 2014;14(1):41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGeachie MJ, Wu AC, Tse SM, Clemmer GL, Sordillo J, Himes BE et al. CTNNA3 and SEMA3D: Promising loci for asthma exacerbation identified through multiple genome-wide association studies. J Allergy Clin Immunol. 2015;136(6):1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.