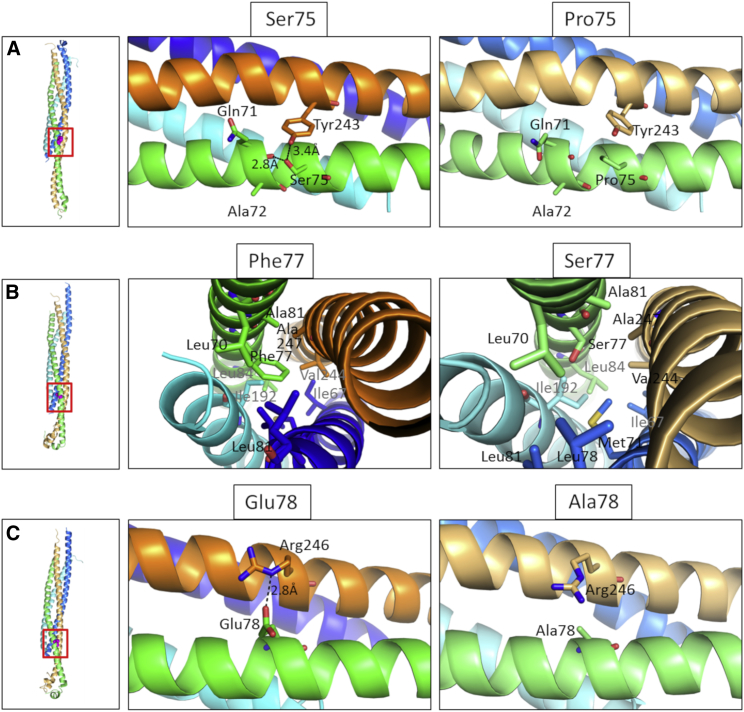
Figure 3.
Molecular Modeling of the Identified De Novo VAMP2 Non-Synonymous Variants
Comparison between the p.Ser75Pro (A), p.Phe77Ser (B), and p.Glu78Ala (C) mutant conformation within the SNARE complex (left panel, red square). The wild-type conformation is shown in the middle panel, and the mutated residues are shown in the right panel. Variant p.Ser75Pro causes the loss of two hydrogen bonds, one interchain between Ser75 of VAMP2 and Tyr243 of STX1A and one intrachain between Ser75 and Gln71; variant p.Phe77Ser introduces a hydrophilic residue in an otherwise hydrophobic region; and variant p.Glu78Ala causes the loss of a hydrogen bond between Glu78 of VAMP2 and Arg246 of STX1A. Modeling of the VAMP2 ectodomain (green for WT, light green for mutants) in complex with STX1A (orange for WT, light orange for mutants) and Snap25 (blue and cyan for WT, marine and aquamarine for mutants); configurations are as seen 100 ns into the molecular dynamic simulation. The complexes were modeled from the humanized 3HD7 complex. Water molecules and ions are not shown.