Fig. 4 |. Integrating capture and long-read sequencing with annotation pipelines.
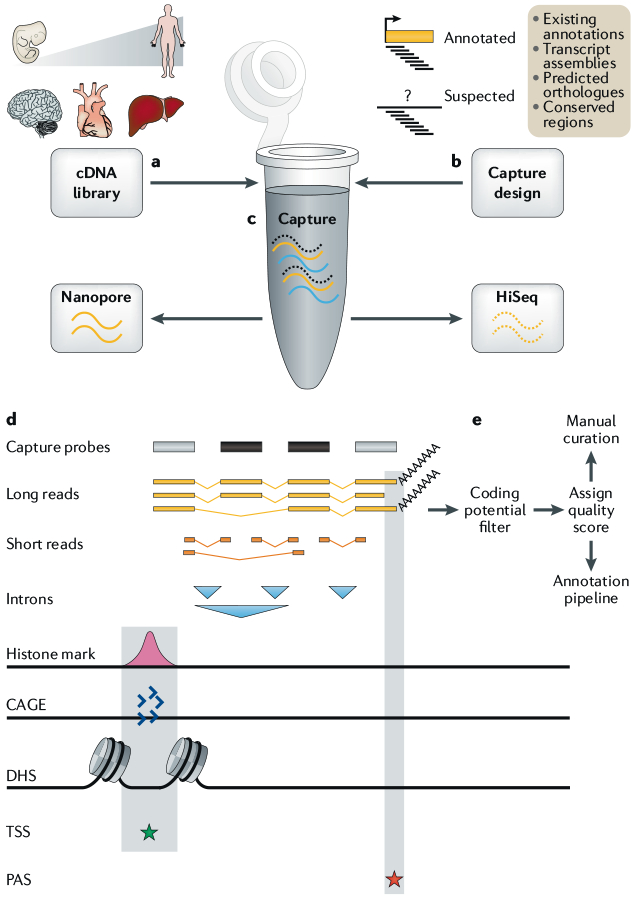
a | Full-length cDNA libraries are prepared from a variety of tissues across the human lifespan. b | Target annotations are prepared from a variety of known and suspected long non-coding RNA (lncRNA) loci and used to design capture probes (black bars). c | Solution-phase oligonucleotide capture is performed, and enriched cDNA libraries are sequenced by long-read nanopore and short-read Illumina technologies. d | The resulting long reads are collapsed to produce non-redundant transcript models. The completeness and accuracy of these models are assessed using various evidence: introns (blue triangles) by short reads; transcription start site (TSS; green star) by promoter histone modifications, cap analysis of gene expression (CAGE) clusters and DNase I hypersensitivity sites (DHSs); and polyadenylation site (PAS; red star) by long-read-encoded poly(A) tails. e | With this information, transcript models are graded for completeness, checked for protein-coding potential and passed to annotators for either direct incorporation into annotation pipelines (for complete models) or further manual curation (incomplete models).
