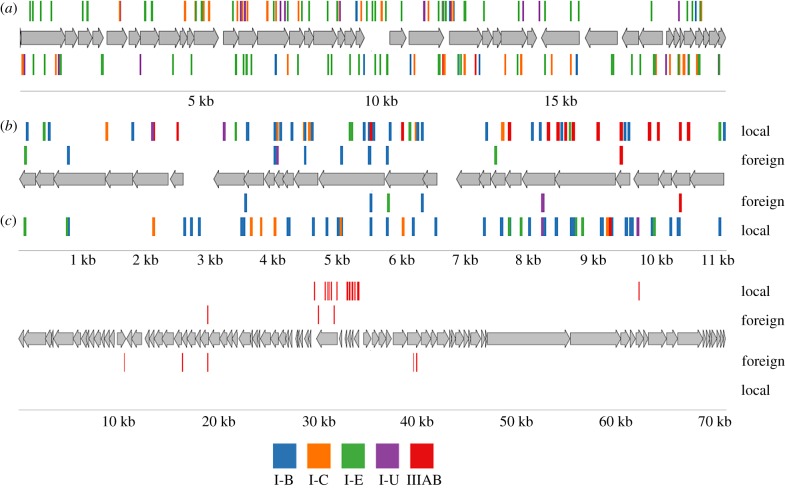
Figure 3.
Mapping of protospacers in the genomes of Thermus phages. The double-stranded DNA genomes of Thermus bacteriophages IN93 (a), phiKo (b) and phiFa (c) are schematically shown. Numbers below indicate genome coordinates, in kilobases. Phage genes are indicated by grey arrows, with arrow directions matching the direction of transcription. Protospacers matching spacers associated with Thermus CRISPR repeats are shown as vertical lines above and below phage genomes. The colour of lines representing protospacers indicates the type of CRISPR-Cas systems to which the matching spacers belong (the colour scheme legend is shown at the bottom of the figure). For phiKo and phiFa, mapped spacers are separated into ‘local’, i.e. found at the site of phage isolation, and ‘foreign’, i.e. found at distant sites.