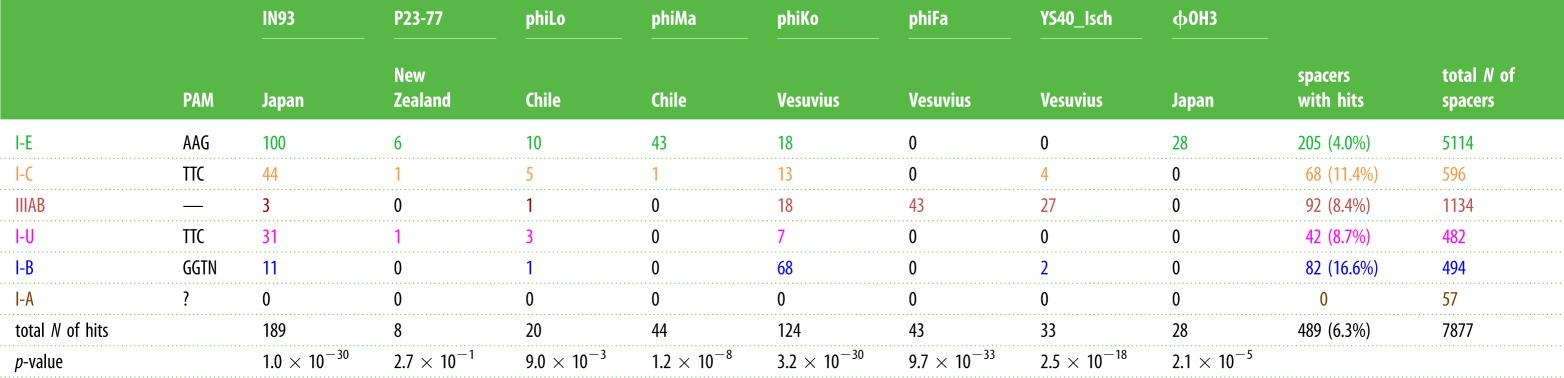
Table 3.
BlastN hits of spacers from different CRISPR-Cas systems. The number of hits for ‘not-unique spacers’, i.e. identical spacers belonging to different CRISPR-Cas system types, is shown. Only hits with greater than 85% identity over entire spacer length are included. The predicted PAMs for each system are presented in the second column (PAM logos are shown in electronic supplementary material, figure S3). Fisher's exact test was used to test for each virus that the number of protospacers depends on the type of CRISPR-Cas system. The resulting p-values are given in the last row.
 |
