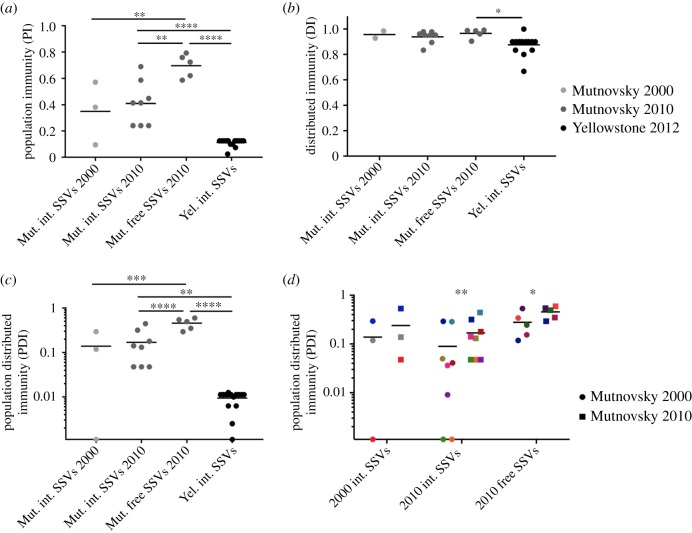
Figure 3.
CRISPR immunity differs by geographical location and with time. (a) The PI to contemporary viruses from the same geographical location and time point (light grey, Mutnovsky 2000; dark grey, Mutnovsky 2010; black, Yellowstone 2012). Spacers with fewer than four mismatches to viral protospacers are considered to provide a strain with immunity if they target a PAM-containing protospacer or come from a strain with a type III CRISPR-Cas system. The average and values for individual viruses are shown. The DI (b) and PDI (c) for the same contemporary virus—S. islandicus interactions shown in (a). Values were compared among virus classes using a one-way ANOVA with Tukey's multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. The PDI (d) for the Mutnovsky S. islandicus populations from 2000 (filled circle) and 2010 (filled square) for the three groups of SSVs identified from Mutnovsky. Matching colours identify a single virus targeted by the two population time points. The immunity to each group of viruses by the two population time points was compared using a paired t-test. (Online version in colour.)