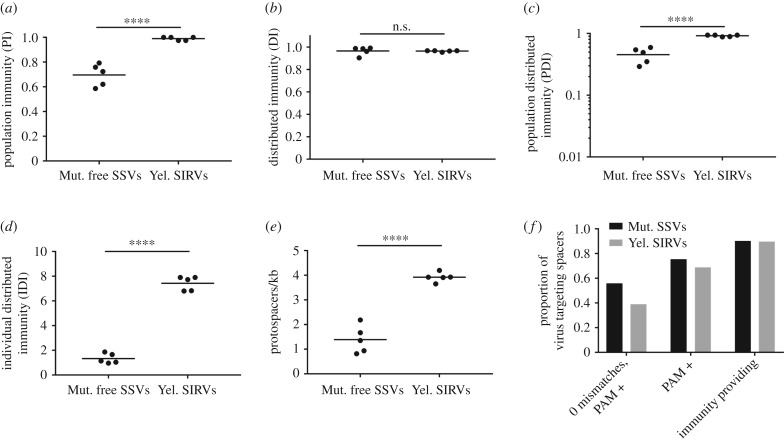
Figure 5.
CRISPR immunity differs by virus type. The PI (a), DI (b) and PDI (c) for the 2010 Mutnovsky population targeting free 2010 Mutnovsky SSVs and the Yellowstone population targeting SIRVs identified from the same Nymph Lake hot springs. Spacers with fewer than four mismatches to viral protospacers are considered to provide a strain with immunity of they target a PAM-containing protospacer or come from a strain with a type III CRISPR-Cas system. (d) The IDI (number of spacers each strain uses to target virus) for the same population–virus interactions as above. (e) For each virus, the number of targeted protospacers per basepair of viral genome. For each measurement, the average and values for individual viruses are shown. The two groups were compared by a two-tailed t-test. n.s., no significance, ***p < 0.001, ****p < 0.0001. (f) The proportion of unique spacers targeting Mutnovsky SSVs or Yellowstone SIRVs with four or fewer mismatches that perfectly match PAM-containing protospacers, match PAM-containing protospacers, or are classified as providing immunity based upon our criteria for strains to possess type III CRISPR-Cas systems to target protospacers lacking PAMs.