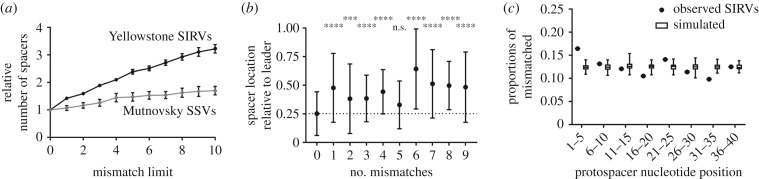
Figure 6.
Viruses evolve in response to targeting by spacers. (a) The relative number of spacers that target PAM-containing protospacers in Yellowstone SIRVs (black) or Mutnovsky SSVs (grey) when the allowable number of mismatches between spacer and protospacer is relaxed. (b) The spacer location relative to the leader end of its CRISPR locus grouped by the number of mismatches to PAM-containing protospacers in Yellowstone SIRVs. The spacer immediately following the CRISPR leader sequence is at 0 and the spacer farthest from the leader sequence is at 1. Error bars indicate standard deviations. The relative location of mismatched spacers was compared with the location of perfectly matching spacers (0 mismatches, dashed line) by one-way ANOVA with Dunnett's multiple comparison test (n.s., no significance, ***p < 0.001, ****p < 0.0001). (c) The locations of all mismatches between spacers and PAM-containing protospacers in Yellowstone SIRVs. Observed data show the proportion of mismatches found within 5 bp windows of the protospacer oriented so the PAM is located upstream of nucleotide position 1. The simulated data represent 2037 mismatches randomly located across protospacers. Whiskers represent the minimum and maximum proportions from 100 simulations.