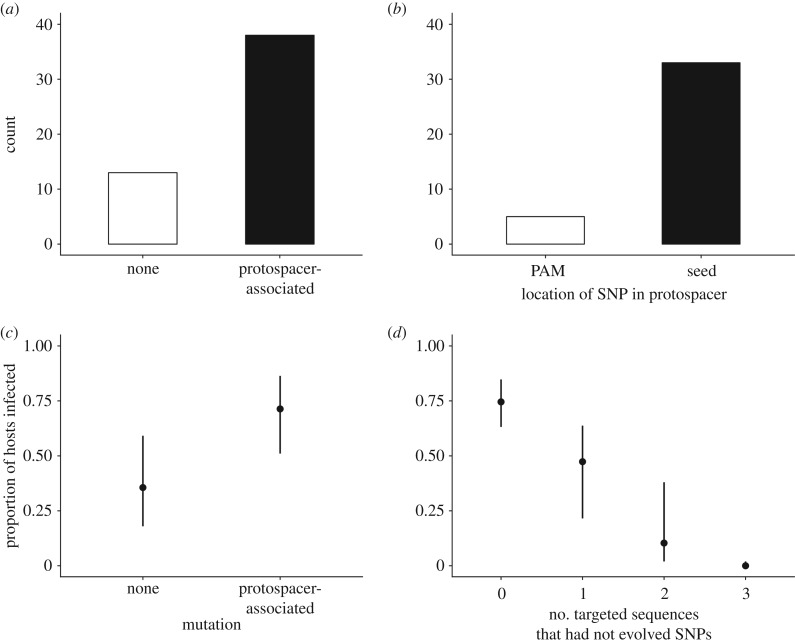
Figure 6.
Protospacer sequence analysis and infectivity patterns. (a) Histogram showing the number of sequenced phage (out of 56) that did not have a detectable mutation, had a ‘random’ mutation outside of the protospacer or had a ‘protospacer-associated’ mutation either in the protospacer-adjacent motif (PAM) or the seed sequence. (b) Number of sequenced phage with protospacer-associated mutations (out of 40) that had a single nucleotide polymorphism (SNP) in either the PAM or seed sequence. (c) Mean proportion of hosts infected by phage that did or did not have a protospacer-associated SNP. The effect of random mutations is not included owing to limited sample size. (d) Mean proportion of hosts infected by phage that had a full or partial match to the host's CRISPR array in terms of the number of targeted protospacer sequences that had not evolved by point mutation. Means and 95% CIs are shown (n = 696).