Abstract
Genetic and environmental factors influence the development of coronary artery disease (CAD). Genetic analyses of families and the population continue to yield important fundamental insights for CAD. For the past four decades, CAD human genetic research focused largely on the study of germline genetic variation in CAD and its risk factors. The first genes associated with CAD were discovered using basic Mendelian principles and pedigree analysis. Mapping of the human genome and advancement in sequencing technology sparked further discovery of novel genetic associations through exome sequencing and genome wide association analysis in increasingly larger populations. While prior work implicated in situ DNA damage as a feature of atherosclerosis, more recently, somatic mutagenesis in and clonal expansion of haematopoietic stem cells was found to influence risk of CAD. Mutations observed for this condition, termed clonal haematopoiesis of indeterminate potential, frequently occur within epigenetic regulator genes (e.g. DNMT3A, TET2, ASXL1, etc.), which are also implicated in leukaemogenesis. Hypercholesterolaemic mice with Tet2 bone marrow deficiency are predisposed to the development of atherosclerosis that may be partly related to inflammatory cytokines. As the genetic basis of CAD expands from the germline to somatic genome, our fundamental understanding of CAD continues to evolve; these new discoveries represent new opportunities for risk prediction and prevention, and a new facet of cardio-oncology.
Keywords: Genetics, Coronary artery disease, Clonal haematopoiesis of indeterminate potential, Somatic mutations
This article is part of the Spotlight Issue on Cardio-oncology.
1. Introduction
Cardiovascular disease is the leading cause of death in the world.1,2 In addition to being the cause of death for over 3.8 million individuals in the member nations of the European Society of Cardiology each year, it accounts for a large share of each nation’s health care system expenditure.3 Much of the mortality and morbidity from cardiovascular disease stems from coronary artery disease (CAD). A deeper understanding of the causes of CAD is essential to guide effective prevention and treatment measures.
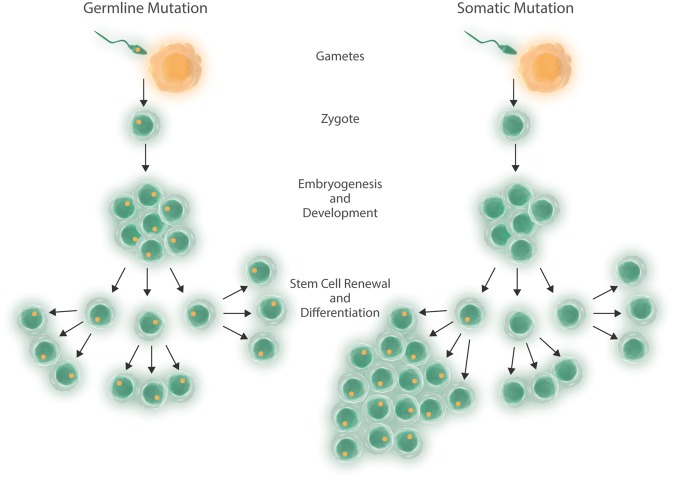
An individual’s genes, environment, and chance dictate the development and severity of CAD and its risk factors.4 The heritability, or trait variability explained by the sum of genetic differences, of CAD is estimated to be 40–60%.5,6 Although the template for germline genomic variation for an individual is established at gametogenesis and fertilization, each mitosis thereafter is subject to the forces of mutagenesis by chance, or biological or environmental influences (Figure 1). In oncology, study of the acquired mutations has been central to understanding the mechanism of tumourigenesis, and although cardio-oncology has emerged as a field due to potential cardiovascular toxicities of oncologic therapies, recent discoveries in clonal haematopoiesis inform new appreciation of common features between cardiovascular diseases and cancer. In this review, we highlight genetic work performed over the past 40 years of largely germline genetic variation to now somatic genetic variation, and their risks for CAD.
Figure 1.
Germline and somatic mutagenesis. In germline mutagenesis, a mutation is transmitted from parent to offspring and all subsequent lineages carry it. In somatic mutagenesis, a mutation occurs in a stem cell or replicating differentiated cell leading to mosaicism, and potentially clonal advantage in replication and survival.
2. Germline variant discovery, Mendelian studies to exome sequencing
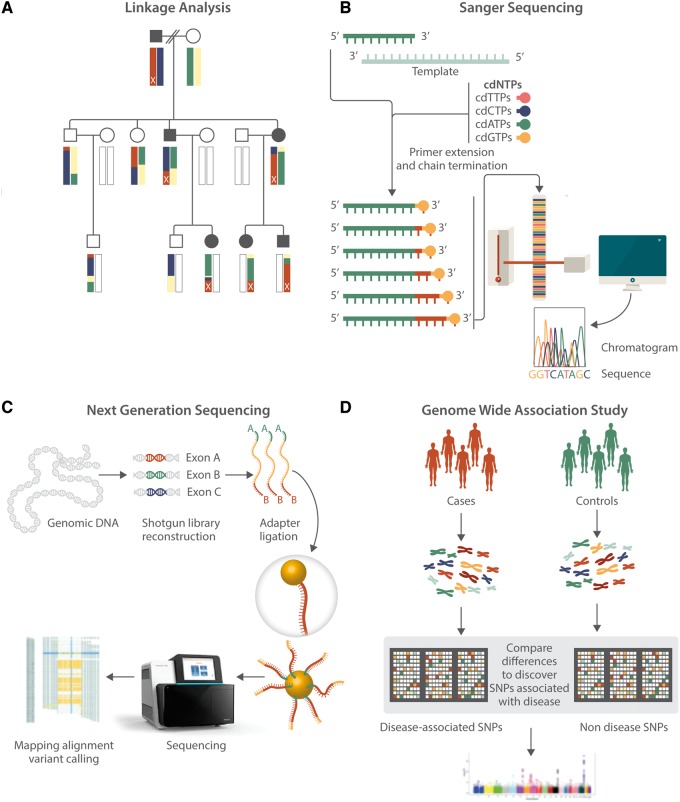
CAD often aggregates in families, particularly when it occurs earlier in life. Early human genetic studies of CAD analysed exceptional families displaying apparent Mendelian segregation of CAD using genetic linkage analysis (Figure 2A). In the first genetic analyses, families with co-segregation of severe hypercholesterolaemia and premature CAD were studied. In one such study, DNA sequence was deduced with cDNA from restriction enzyme digestion patterns and blot hybridization; a 5 kb deletion in the gene encoding for the low-density lipoprotein (LDL) receptor (i.e. LDLR) deficient in cell membrane adherence and LDL clearance was discovered.7,8 Advancement in genetic mapping and development of Sanger sequencing technology led to further understanding of the sequence and variation in LDLR and other genes involved in syndromes of familial dyslipidaemia and myocardial infarction (MI) (Figure 2B).9–11 In the study of families with apparent autosomal dominant severe hypercholesterolaemia but without LDLR mutations, mutations (later characterized to be gain-of-function) in the gene PCSK9 were implicated through positional cloning and parametric linkage analyses.12 At the population-level, loss-of-function mutations in PCSK9 were associated with marked reductions in LDL cholesterol concentration and subsequent reduction in risk of CAD in both European and African Americans.13
Figure 2.
Advances in genetic analysis methodology. (A) Linkage analysis is a statistical method that tracks and correlates hereditary phenotype transmission with genetic loci relying on the consequences of genetic recombination between two autosomal chromosomes during meiosis. Finer resolution mapping is possible with larger pedigrees. This schematic depicts an autosomal dominant trait originating from the terminal chromosomal locus in the pedigree father (marked x) and being transmitted to a granddaughter via an affected son and to a grandson and granddaughter via an affected daughter. (B) Sanger sequencing is used to determine the DNA sequence for a locus. Repetitive polymerase chain replication is performed in which a constitutively replicated sequence fragment is randomly terminated with a labelled chain-terminating dideoxynucleotide. Several sequencing fragments of varying lengths and end nucleotides emerge and are separated based on length. The attached base for each length is imaged to yield a sequence of bases. (C) Next generation sequencing refers to a group of methods of large-scale, parallel sequencing using Sanger sequencing principles. This involves fragmenting genomic DNA, attaching adaptors to the newly formed DNA fragments, attaching these adaptors to a solid surface for sequencing, amplifying the fragmented DNA strands in parallel, enriching desired segments, sequencing in parallel using different methods, aligning the sequence reads that result from the pooled results to known genomic reference sequence, and examining the resulting DNA variants. (D) Genome wide association (GWA) involves using large genotyping arrays containing SNPs, each representative of a locus of bases that are generally inherited together due to linkage disequilibrium. DNA from cohorts of individuals affected and unaffected by a trait are genotyped with these arrays and the comparative differences in prevalence of certain mutations are used to statistically determine associations of polymorphisms at a certain locus with the trait of interest.
Advances in next-generation sequencing technology now permit sequencing of the exome (i.e. the full protein-coding portion of the genome) or even the whole genome itself (Figure 2C). Whole exome analysis also facilities gene discovery within exceptional families. For example, exome sequencing of a family with apparent Mendelian combined hypolipidaemia identified causative nonsense mutations in ANGPTL3, an inhibitor of lipoprotein lipase and endothelial lipase.14 Furthermore, advances now permit scalable application for gene discovery in large studies (e.g. case-control, cohort, etc). Exome sequencing of 9793 individuals of patients with early-onset MI and MI-free controls verified that variants in LDLR associated with 4.2–13-fold odds for MI, and showed that variants in APOA5 associated with 2.2-fold odds of MI, each also with associations of hypercholesterolaemia and hypertriglyceridaemia, respectively.15 A cohort study with exome sequences of 3734 individuals observed that loss-of-function mutations in APOC3 were associated with lower levels of triglycerides and reduced risk of CAD.16,17
3. Germline variant discovery, the genome wide association study
While the analyses of rare alleles and their influences on cardiovascular traits can lead to broad insights about cardiovascular disease, a larger fraction of the heritability of common traits such as CAD appears to be explained by common genetic variation. Genome-wide association studies (GWAS) catalogue common coding and non-coding variation across the human genome through single nucleotide polymorphism (SNP) arrays and associate these variants with quantitative or discrete traits in population-based analyses (Figure 2D).
Over a decade ago, the first GWAS of CAD/MI showed that the top association was at the 9p21 locus, a genomic region not previously implicated in cardiovascular disease.18–20 As GWA studies increase in size and ethnic diversity, novel loci continue to be discovered.21–27 The most recent and largest meta-analyses identified 64 new loci implicated in CAD, bringing estimated total to over 150 loci reaching genome wide significance.24,28,29 Additionally, international collaborations identified numerous associations with risk factors implicated in CAD, notably in hypertension,30,31 lipids,32 and Type 2 diabetes.33–35 At the individual-level, risk alleles can be summed as a weighted polygenic risk score to quantify genetic predisposition to CAD as well as its risk factors.36–38
Although associations are abundant, explaining the true disease mechanism of GWAS signals at regions without previously implicated Mendelian genes remains challenging. Disentangling the causal variant itself can be challenging as the lead significant variant with the strongest statistical signal is correlated with several other variants in linkage disequilibrium. Additionally, many of the implicated variants are in non-protein-coding regions, making hypothesis-driven candidate testing challenging. Musunuru et al.39 reported one common, non-coding GWAS-implicated variant at the chromosome 1p13 locus to be involved in a novel regulatory pathway, where altered transcription factor binding leads to altered hepatic expression of SORT1. This gene is involved in pre-secretory degradation of very low-density lipoprotein (VLDL) cholesterol and leads to altered LDL cholesterol and VLDL cholesterol levels, thereby influencing risk for CAD.
4. In situ DNA damage and atherogenesis
Separate from the analysis of germline genetic variation, various investigational methods in model systems previously linked acquired damage to DNA and repair defects within endothelial cells and cholesterol plaques with atherogenesis.40 The process of atherosclerosis is mediated by damage to endothelium, deposition of cholesterol, activation of inflammation and fibroproliferation.41 Cholesterol bound to lipoprotein particles in the bloodstream attach to intima and aggregate to form the beginnings of fatty streaks. These particles undergo chemical modifications in the setting of oxidative stress.42 Deletions or additions of whole or parts of chromosomes as well as loss-of-heterozygosity (LOH), strand breaks, base pair modification, and microsatellite instability (MSI) have been observed in the plaque environment. The phenomenon of vascular aging is thought to occur as a result of exposure to reactive oxygen species (ROS) and resultant genomic instability, which subsequently affects normal cellular function within atheroma.43
Oxidative damage to mitochondrial genes promotes the generation of ROS in vascular tissues and atherosclerotic lesions. Atherosclerotic plaques are enriched for markers of immunoreactivity against oxidized DNA with concomitant local up-regulation of DNA repair systems.44 Mitochondrial DNA damage correlates with atherosclerotic extent in humans and in mice.45,46 Furthermore, mitochondrial damage is widespread in cells involved in atherosclerosis and may promote lesion development.47
MSI and LOH are common genomic alterations observed in cellular nuclei within atherosclerotic plaques. Microsatellites are short, repeated sequences of DNA, and instability in their inheritance occurs when the number of repeats changes with erroneous replication. LOH is a phenomenon that occurs when a heterozygote loses a wild-type allele through mutagenesis or non-disjunction. Several human studies of atherosclerotic plaques showed greater burden of LOH and MSI in atheromatous plaques compared with unaffected vascular tissue within the same individual.48 MSI is particularly enriched at proto-oncogene regulators and genes central to signal transduction in vascular repair and healing, implying a causal mechanism of disordered proliferation and transformation of disease-related smooth muscle cells with atherosclerosis.49,50 Furthermore, such mutagenesis may facilitate selection since large clonal chromosomal abnormalities are more commonly observed in the smooth muscle cells from unstable atherosclerotic plaques than in stable plaques.51 However, it remains unclear if observed plaque characteristic differences are causes or effects of the observed genetic instability.
Telomere dysfunction may also lead to genomic instability and to atherogenesis. Telomere shortening in blood leucocytes is associated with metabolic syndrome, diabetes mellitus, and CAD.52–54 Within vascular smooth muscle cells, accelerated telomere attrition and replicative senescence is associated with upstream angiotensin II-induced ROS-mediated DNA damage.55 Similarly, it remains unclear if this is a cause or consequence of atherogenesis. Nevertheless, statin use is correlated with accelerated DNA repair in vascular smooth muscle cells, reduced telomere shortening, and reduce vascular smooth muscle cell death.56
5. Somatic mutations in blood cells conferring clonal advantage
In addition to the passive deposition and transformation of cholesterol that occurs in the endothelium, monocytes play a central role in the formation of atheroma.42 Somatic mutations can occur within the monocytes interacting with vascular tissues and may separately influence atherosclerosis.
Leucocytes are recruited to the intima through expression of extracellular proteins in the cell adhesion molecule, integrin, and selectin families. Monocytes subsequently accumulate lipids via scavenger receptors and subsequently transform into foam cells. These cells thereafter produce further inflammatory mediators as well as oxidant species that contribute to progression of atherosclerotic lesions. The vascular smooth muscle cells then rapidly multiply, produce extracellular matrix molecules, and foster calcification, leading to maturation and evolution of the atherosclerotic plaque.42
Haematopoietic stem cells continuously regenerate providing an ideal opportunity for selection and proliferation in the setting of mutagenesis (Figure 1). DNA from whole blood for germline DNA analysis is extracted from peripheral mononuclear cells; separate bioinformatics software can be used to distinguish the presence of acquired mutations of appreciable allele frequency (indicating clonal expansion).
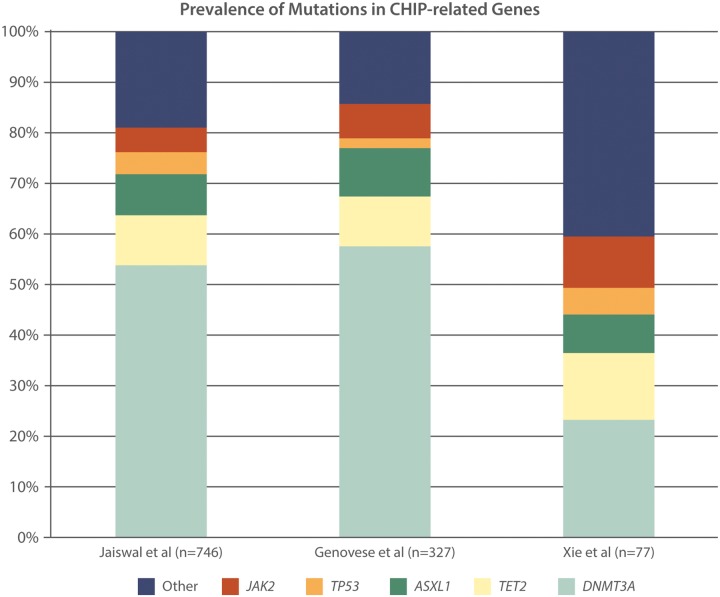
Xie et al.57 studied 2728 individuals with non-haematologic malignancy from The Cancer Genome Atlas and identified 77 somatic mutations in cancer genes in blood cells from whole exome sequencing. The majority of these mutations are commonly associated with haematologic malignancies, with nine loci that are recurrently mutated (DNMT3A, TET2, JAK2, ASXL1, TP53, GNAS, PPM1D, BCORL1, and SF3B1, Figure 2).
Further studies among individuals without prior malignancy demonstrated that clonal haematopoiesis with pre-malignant somatic mutations is associated with increased risk of haematologic cancer and death. From the analysis of 12 380 blood-derived whole exome sequences, Genovese et al.58 identified 3111 somatic mutations in white blood cells based on the presence of unusual allelic fractions dispersed across the exome. There was similarly disproportionate enrichment of disruptive mutations in the genes implicated in haematologic malignancy, namely DNMT3A, TET2, and ASLX1 (Figure 3). Prevalence was associated with age: 1% of individuals younger than age 50 were carriers and 10% of individuals older than age 65 were carriers. Furthermore, clonal haematopoiesis was associated with the development of haematologic malignancy, conferring a hazard ratio of 12.9 [95% confidence interval (CI) 5.8–28.7].58
Figure 3.
Prevalence of the most common mutations implicated in clonal haematopoiesis of indeterminate potential (CHIP) in three seminal studies.
Jaiswal et al. also report an age-related increase in observed frequency of clonal somatic mutations. In this blood-derived whole exome sequencing study of 17 182 individuals, they also reported that the majority of SNPs that were found in the same leukaemogenic genes (DNMT3A, TET2, and ASLX1, Figure 3). Not only was carrying these mutations associated with an increased risk of haematologic cancer but also all-cause mortality (hazard ratio 1.4, 95% CI 1.1–1.8). Exploratory analyses suggested an association with CAD (hazard ratio 2.0, 95% CI 1.2–3.4) and ischaemic stroke (hazard ratio 2.6, 95% CI 1.4–4.8) despite accounting for conventional risk factors.59
Notably, clonal haematopoiesis as detected by the presence of a pre-malignant mutation of variant allele fraction at least 2% in the absence of overt malignancy or other benign haematological conditions is termed ‘clonal haematopoiesis of indeterminate potential’ (CHIP).60–62
Recently, a whole genome sequencing study in Iceland demonstrated that clonal development has a heritable basis. In this study, a common 8-bp deletion in an intron of the telomerase reverse transcriptase gene (TERT) increases risk for developing clonal haematopoiesis, though the mechanism currently remains unclear.63
6. Clonal haematopoiesis of indeterminate potential and atherosclerosis
To examine the role of CHIP in CAD more closely, we exome sequenced 4726 participants with CAD and 3529 without known CAD, as defined by history of MI or revascularization from a total of four cohorts. As before, most mutations among those with CHIP were observed in DNMT3A, TET2, and ASXL1. CHIP carriers were found to have 1.9 times the risk of CAD when compared with non-carriers (95% CI 1.4–2.7; P < 0.001). Remarkably, despite being an age-associated condition, younger carriers were at 4.0 times the risk of early-onset MI when compared with non-carriers (95% CI 2.4–6.7; P < 0.001). Narrowing the pool of mutations to just DNMT3A, TET2, and ASXL1 revealed that participants with these mutations had 1.7 to 2.0 times the risk of CAD compared with those without these mutations, and the p.V617F mutation in JAK2 was noted to confer 12.1 times the risk. Similar results were found with early-onset MI. Furthermore, participants with CHIP mutations were noted to have 3.3 times higher amount of coronary artery calcification as non-carriers. Lastly, increased CHIP allele fraction, indicating a larger clone size, correlated with greater effect size as well. Patients with CHIP variant allele fraction >10% had 2.2 times the risk of having CAD, compared with 1.4 times the risk seen in those with variant allele fraction of less than 10%.64
To further assess for causality, we investigated the role of TET2, a gene involved in DNA methylation and transcription regulation, further in a mouse model of hypercholesterolaemia and atherosclerosis. Ldlr−/− mice were irradiated and transplanted with bone marrows of Tet2−/− mice or Tet2+/+ mice. The hypercholesterolaemic mice with transplanted Tet2 deficient bone marrow had twice the size of aortic root median atherosclerotic lesion size compared with hypercholesterolaemic mice transplanted with wild-type bone marrow. An intermediate phenotype was observed with Tet2+/− bone marrow Ldlr−/− mice.
To more specifically assess the consequence of knocking out Tet2 on macrophage function, the above experiment was repeated with bone marrow from mice with just myeloid lineage Tet2 knock out constructed using a Cre-Lox system; the lesion sizes in these mice were still observed to be 1.7 times as large as those of controls at 10 weeks. Ex vivo analysis showed that inflammatory cytokines and chemokines were up-regulated in Tet2−/− macrophages vs. Tet2+/+ macrophages. The human cytokine analogue of CXC (i.e. IL-8) was subsequently found to be higher in humans with TET2-associated CHIP compared with unaffected individuals.64
Fuster et al. created a model of competitive bone marrow transplantation with 10% Tet2 deficient HSPCs and 90% Tet2 wild type in place of irradiated bone marrows of Ldlr−/− mice. They demonstrated that the Tet2 bone marrow deficiency had rapid clonal expansion without affecting cell counts, weight, spleen weight, glucose, or cholesterol levels. They further demonstrated that hypercholesterolaemic mice with Tet2 bone marrow deficiency had larger aortic root plaque burden compared with hypercholesterolaemic mice with transplanted wild-type bone marrow. In vitro assays of macrophage activation showed significant alteration in inflammatory cytokine and chemokine expression between Tet2−/− and Tet2+/+ macrophages. Through various provocations, they showed that IL-1B may particularly link Tet2 deficiency with atherosclerosis (Figure 4). Furthermore, inhibition of the NLRP3 inflammasome, an activator of IL-1B, mitigated atherosclerosis plaque size in the setting of Tet2−/− bone marrow in a murine model.65
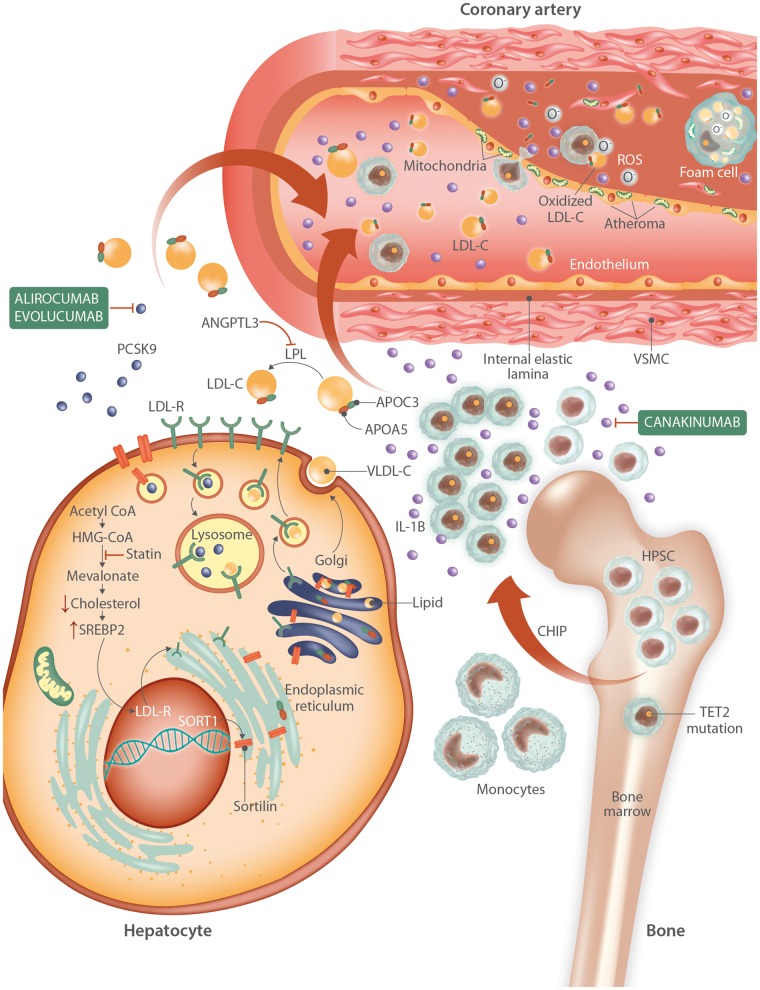
Figure 4.
Interplay of well-known germline and somatic mutations in atherogenesis. In the liver, cholesterol biosynthesis begins with acetyl-CoA, and intracellular cholesterol levels are regulated with assistance the SREBP2 pathway. Statins interfere with cholesterol synthesis by inhibiting HMG-CoA reductase, leading to a drop in intracellular cholesterol levels, synthesis of more LDL receptors, and increased uptake of LDL cholesterol from the circulation. PCSK9 molecules help regulate the number of LDL receptors on the cell surface by aiding in their uptake from the cell membrane and transport to the lysosome for degradation. Evolocumab and alirocumab inhibit the activity of PCSK9, allowing more LDLR-mediated uptake of serum cholesterol. Sortilin expression, mediated by enhancers at the chromosome 1p13 locus, promotes release of mature VLDL particles into the circulation, PCSK9 secretion, and macrophage lipid accumulation leading to foam cell formation. In the circulation, various forms of lipoproteins combined with lipid molecules transport their contents throughout the body. APOA5 and APOC3 on VLDL particles participate in triglyceride metabolism and inhibit lipoprotein lipase (LPL), respectively. ANGPTL3 also inhibits LPL, leading to increased circulating levels of cholesterol and triglycerides, which are deposited in the endothelium of the vasculature. In the bone marrow, somatic mutations in TET2 lead to hyperproliferative advantage for a subset of haematopoietic pluripotent stem cells (HPSCs), leading to clonal haematopoiesis of indeterminate potential (CHIP). The clonal monocytes that result from further replication produce an abundance of IL-1 beta, which promotes further inflammatory cascades and is in part inhibited by canakinumab. These macrophages adhere to the lipid-rich endothelium and traverse it. Within the vessel wall, mitochondrial dysfunction and generation of ROS leads to oxidation of LDL, generation of more inflammatory cytokines, and further damage to surrounding cells. Macrophages consume this oxidized LDL to become foam cells. Vascular smooth muscle cells (VSMCs) proliferate, damaged cells apoptose, and the atheroma continues to grow.
7. Clonal haematopoiesis with chromosomal alterations
Apart from SNPs (mutations typically observed in CHIP) leading to cell’s increased proliferation, age-related large clonal chromosomal alterations have also been observed in blood cells. Jacobs et al.66 studied 31 717 cancer cases and 26 136 cancer-free controls from 13 GWAS and found significant aneuploidy or copy neutral LOH in a subset of clones. Like with CHIP, age remains the strongest predictor of mosaic chromosomal abnormalities. Frequency increased from 0.23% for cancer free individuals under age 50 years to 1.91% in individuals between age 75 and 79 years. Furthermore, these abnormalities were noted to be more common in those with solid tumours.66
Although external and stochastic factors may influence the development of somatic large chromosomal alterations, Loh et al.67 recently demonstrated that germline genetic variation also plays a role. They studied 8342 mosaic chromosomal alterations ranging in size from 50 kb to 249 Mb in blood DNA from 151 202 individuals from UK Biobank. Using phase-based computational techniques, they identified three germline genomic loci (MPL, TM2D3/TARSL2, and FRA10B) associated with clonal chromosomal mosaicism in blood cells. They demonstrated that chromosomal alterations of trisomy 12, 13q, and +3/3q+ correlated strongly with incidence of CLL after 1 year; similarly 9p abnormalities were linked with risk of myeloproliferative neoplasms. Additionally, this condition was strongly linked with increased overall mortality.67
Despite these multiple associations of mosaic chromosomal abnormalities identified with cancers, less is known regarding associations with cardiovascular disease.67 Bonnefond et al.68 previously studied the role of large chromosomal clonal mosaic events and Type 2 diabetes. They examined chromosomal alterations in 7659 individuals, 2208 of which had diabetes, using DNA arrays and reported a significant association between the presence of alterations and diabetes (odds ratio 5.3, P 5.1 × 10−5). In secondary analyses among diabetics, they noted that those with large chromosomal clonal mosaicism had a higher prevalence of microvascular or macrovascular complications compared with those without these alterations (19/26 vs. 810/2182, P 7.7 × 10−4). It is currently unclear whether such clonal chromosomal abnormalities are the cause or consequence of Type 2 diabetes.68
8. Future directions
With ongoing evolution in our understanding of the complex genetic underpinnings of CAD, more questions and opportunities continue to arise. Just as understanding of principles of tumourigenesis have helped reveal novel mechanisms of atherogenesis, the discovery techniques and therapeutics from the genetic study of CAD can be applied more broadly to cardio-oncology.
8.1 Germline directions
8.1.1 Discovery
With advancement in technology and lower costs, large-scale genetic sequencing and genotyping is becoming more ubiquitous across research, clinical, and direct-to-consumer settings.69 Large consortia of researchers are comprehensively cataloguing the human germline genome and a multitude of somatic mutations responsible for causing cancer.70–72 In pursuit of precision medicine, many national health systems and academic medical centres are amassing hundreds of thousands of samples of blood and tissue from patients.73–76 With increasing sample sizes and increasing diversity, new genes and genetic variants related to cardiovascular disease and susceptibility to cardiovascular toxicities from oncologic therapies will be discovered.
8.1.2 Biological elucidation
GWAS already identified hundreds of new SNPs related to CAD and its risk factors in large populations. Most associated SNPs are non-coding (intergenic or intronic) and fall in regions of linkage disequilibrium extending tens of thousands of bases. These regions often contain many protein-coding genes. A major challenge at each GWAS locus is the identification of the culprit variant and gene as well as the mechanistic relationship between the two responsible for the associated outcome. Targeted sequencing has been applied with limited success in identifying novel loci within these regions.77 Scalable, systematic approaches robust to diverse biological processes are urgently needed and require interdisciplinary collaboration.78
8.1.3 Risk prediction
With the ongoing accumulation of knowledge of SNP and disease association, polygenic risk scores for age-agnostic risk prediction are increasingly improving. Polygenic risk scores can be applied to identify individuals at future risk for virtually any heritable condition, including CAD, orthogonal to conventional approaches.79 Similarly, variation in biomarkers and SNPs have been used to predict risk of anthracycline-induced cardiotoxicity risk, and this can further be expanded to other oncologic therapies involved in cardiovascular disease development.80,81 With respect to CAD, genetic predisposition may also influence anticipated response to preventive strategies.82 Continued study of the genetic basis of CAD in non-Europeans will improve polygenic risk prediction in these groups. Furthermore, prospective clinical trials testing the clinical efficacy of polygenic risk scoring are required.
8.2 Somatic directions
8.2.1 Discovery
By definition, CHIP requires the presence of leukaemogenic mutation in a previously implicated gene. However, many individuals have evidence of clonal haematopoiesis without driver mutations in known genes offering opportunities to discover newly implicated genes.59 Additionally, while our understanding of the relationship between CHIP and CAD continues to progress, the clinical significance of clonal chromosomal mosaicism with respect to CAD is still less clear. Clonal selection and associated oncogenic somatic mutations were recently observed across tissue types in asymptomatic individuals83 but whether this phenomenon is associated with additional risk for cardiovascular disease is currently not known. Furthermore, various chemotherapies, including platinum-based84,85 and antimetabolite chemotherapies,86 growth factor inhibitors,87 and radiation therapy88,89 are linked to increased incidence of vascular ischaemia but the contribution of DNA damage and mutagenesis is unknown.
8.2.2 Biological elucidation
Recent studies highlighted the role of TET2 in atherosclerosis. However, the putative atherogenic mechanisms from other CHIP genes warrants further investigation. DNMT3A, a DNA methyltransferase and the most commonly mutated of the CHIP genes, is involved in de novo DNA methylation and epigenetic regulation in development, and mutations in the gene have been liked to acute myeloid leukaemia.90,91 Inflammation-related pathways are implicated in TET2 downstream signalling, however, it remains to be discovered if any other processes are found to contribute to CHIP-related atherogenesis.
Despite being a strong risk factor for age-related diseases, a minority of those with CHIP develops haematologic malignancy or CAD. Discovery and characterization of the predictors of CHIP-related clinical consequences may help clarify mechanistic relationships. For example, whole blood or single cell RNA-seq complemented by DNA-seq assessed longitudinally may help elucidate the biological processes linked to driver mutation onset, clonal expansion, and ultimate clinical outcome. Additional biochemical profiling may provide additional insights. To verify alterations are not reflective of reverse confounding, causal inference analyses using conventional and genetic epidemiology approaches and experimental studies in model systems will likely be necessary.
Furthermore, although we identify CHIP based on a set of commonly identified mutations in genes involved in haematologic malignancies, the clones that these mutated transcriptional regulators lead to have a host of passenger variants that also become clonal by proxy. Whether such passenger variants, independent from clone size, influence CAD risk is currently unknown.
Further knowledge of mechanistic insight of cytokines in CAD as seen in CHIP may also lead to broader connections of inflammation in atherosclerosis. This has the potential to further refine our understanding of the accelerated atherosclerosis in patients with human immunodeficiency virus (HIV) infection and autoimmune conditions such as psoriasis, where inflammation is presumed to be a unique driving force of atherosclerosis.
8.2.3 Risk prediction
It is now clear that the genetic basis for CAD extends from the germline to somatic genome. Currently, clinical genetic testing for CAD is largely restricted to gene panel testing for familial hypercholesterolaemia. Our work demonstrates that, at least among those with early-onset MI, the prevalence of CHIP and MI risk conferred is similar to that of familial hypercholesterolaemia.
Separately, array-based technologies are now implemented in the calculation of polygenic risk scoring to quantify heritable risk of CAD. However, this approach neglects the contribution of well-established Mendelian risk mutations and the newly-recognized influence of rare somatic clonal mutations. Whole genome sequencing offers the opportunity to most comprehensively quantify CAD genetic risk across germline and somatic variant classes. With technological advances and reductions in cost, implementation and interpretation of whole genome sequences for CAD risk will become increasingly feasible.
8.3 Therapeutics
8.3.1 Germline
The discovery of genes implicated in CAD has prompted the development of new therapeutic agents. For example, PCSK9 inhibitors reduce LDL cholesterol, and consequently, cardiovascular disease events.92,93 Agents targeting other recently discovered targets such as ANGPTL3 are in active clinical trials.94,95 Better mechanistic understanding of other germline targets may further identify orthogonal therapeutic strategies.
8.3.2 Somatic
Knowledge of CHIP carrier status may influence statin decisions as other similarly associated ‘risk-enhancing’ factors.96 Whether those with CHIP have a different relative benefit from statins vs. those who do not requires further study.
Screening for genetic mutations in tumour samples is increasingly being used in oncology to more appropriately match patients to therapies and guide clinical trials.97–99 With further knowledge of CHIP-related and other potential somatic mutations involved in atherosclerosis, a similar testing panel may help guide a personalized approach to managing a particular individual’s CAD risk.
Vitamin C therapy was recently shown to mimic TET2 restoration in otherwise deficient cells undergoing aberrant self-renewal via promoting DNA demethylation, differentiation, and cell death.100 In CHIP clones where TET2 is the driver, vitamin C therapy may also help curtail the implicated downstream cardiovascular pathology.
Since IL-1B is believed to be a key contributor to TET2 deficiency-driven atherosclerosis, inhibition of IL-1B may be particularly efficacious for those with CHIP. In the CANTOS trial, canakinumab, an inhibitor of IL-1B, was shown to reduce recurrent cardiovascular risk among 10 061 patients with prior MI and elevated high-sensitivity C-reactive protein.101 Preliminary analyses within the CANTOS trial suggests that individuals with CHIP and TET2 mutations, experience a greater relative clinical benefit from canakinumab.102
Finally, genomic editing research in the past decade has led to significant breakthroughs leading to more efficient and accurate editing methods. Use of platforms such as CRISPR/cas9 paired with the appropriate delivery apparatus to implicated somatic cells may be curative in diseases stemming from somatic mutations, such as CHIP and atherosclerosis.103
9. Conclusions
Studying genetic variation and its association with CAD permits unbiased evaluation of CAD risk, development and progression, and treatment approaches. Germline associations identified through GWAS and next-generation sequencing continue to yield important insights. More recently, age-related clonal haematopoiesis with pre-leukaemic mutations was shown to influence CAD broadening the genetic basis of CAD to the somatic genome. Efforts to improve understanding of the genetic contribution to CAD can continue to advance our understanding of CAD.
Conflict of interest: P.N. reports grants from Amarin, Amgen, and Boston Scientific, and consulting income from Apple.
Funding
P.N. is supported by a grant from the National Heart, Lung, and Blood Institute (K08HL140203) and from a Massachusetts General Hospital Hassenfeld Scholar award.
References
- 1. Nowbar AN, Howard JP, Finegold JA, Asaria P, Francis DP.. 2014 global geographic analysis of mortality from ischaemic heart disease by country, age and income: statistics from World Health Organisation and United Nations. Int J Cardiol 2014;174:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy SL, Xu J, Kochanek KD, Curtin SC, Arias E.. Deaths: final data for 2015. Natl Vital Stat Rep 2017;66:1–75. [PubMed] [Google Scholar]
- 3. Group AW, Timmis A, Townsend N, Gale C, Grobbee R, Maniadakis N, Flather M, Wilkins E, Wright L, Vos R.. European Society of Cardiology: cardiovascular disease statistics 2017. Eur Heart J 2017;39:508–579. [DOI] [PubMed] [Google Scholar]
- 4. Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, Fuster V, Boerwinkle E, Melander O, Orho-Melander M, Ridker PM, Kathiresan S.. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med 2016;375:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer M, Broeckel U, Holmer S, Baessler A, Hengstenberg C, Mayer B, Erdmann J, Klein G, Riegger G, Jacob HJ, Schunkert H.. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation 2005;111:855–862. [DOI] [PubMed] [Google Scholar]
- 6. Mayer B, Erdmann J, Schunkert H.. Genetics and heritability of coronary artery disease and myocardial infarction. Clin Res Cardiol 2007;96:1–7. [DOI] [PubMed] [Google Scholar]
- 7. Brown MS, Goldstein JL.. A receptor-mediated pathway for cholesterol homeostasis. Science 1986;232:34–47. [DOI] [PubMed] [Google Scholar]
- 8. Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW.. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science 1985;227:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH.. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 2000;290:1771–1775. [DOI] [PubMed] [Google Scholar]
- 10. Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N.. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science 2001;292:1394–1398. [DOI] [PubMed] [Google Scholar]
- 11. Soria LF, Ludwig EH, Clarke H, Vega GL, Grundy SM, McCarthy BJ.. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA 1989;86:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C.. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 2003;34:154–156. [DOI] [PubMed] [Google Scholar]
- 13. Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH.. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 2006;354:1264–1272. [DOI] [PubMed] [Google Scholar]
- 14. Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S.. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med 2010;363:2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, Guella I, Asselta R, Lange LA, Peloso GM, Auer PL, Girelli D, Martinelli N, Farlow DN, DePristo MA, Roberts R, Stewart AF, Saleheen D, Danesh J, Epstein SE, Sivapalaratnam S, Hovingh GK, Kastelein JJ, Samani NJ, Schunkert H, Erdmann J, Shah SH, Kraus WE, Davies R, Nikpay M, Johansen CT, Wang J, Hegele RA, Hechter E, Marz W, Kleber ME, Huang J, Johnson AD, Li M, Burke GL, Gross M, Liu Y, Assimes TL, Heiss G, Lange EM, Folsom AR, Taylor HA, Olivieri O, Hamsten A, Clarke R, Reilly DF, Yin W, Rivas MA, Donnelly P, Rossouw JE, Psaty BM, Herrington DM, Wilson JG, Rich SS, Bamshad MJ, Tracy RP, Cupples LA, Rader DJ, Reilly MP, Spertus JA, Cresci S, Hartiala J, Tang WH, Hazen SL, Allayee H, Reiner AP, Carlson CS, Kooperberg C, Jackson RD, Boerwinkle E, Lander ES, Schwartz SM, Siscovick DS, McPherson R, Tybjaerg-Hansen A, Abecasis GR, Watkins H, Nickerson DA, Ardissino D, Sunyaev SR, O'Donnell CJ, Altshuler D, Gabriel S, Kathiresan S.. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A.. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med 2014;371:32–41. [DOI] [PubMed] [Google Scholar]
- 17.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute, Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, Lu Y, Tang ZZ, Zhang H, Hindy G, Masca N, Stirrups K, Kanoni S, Do R, Jun G, Hu Y, Kang HM, Xue C, Goel A, Farrall M, Duga S, Merlini PA, Asselta R, Girelli D, Olivieri O, Martinelli N, Yin W, Reilly D, Speliotes E, Fox CS, Hveem K, Holmen OL, Nikpay M, Farlow DN, Assimes TL, Franceschini N, Robinson J, North KE, Martin LW, DePristo M, Gupta N, Escher SA, Jansson JH, Van Zuydam N, Palmer CN, Wareham N, Koch W, Meitinger T, Peters A, Lieb W, Erbel R, Konig IR, Kruppa J, Degenhardt F, Gottesman O, Bottinger EP, O'Donnell CJ, Psaty BM, Ballantyne CM, Abecasis G, Ordovas JM, Melander O, Watkins H, Orho-Melander M, Ardissino D, Loos RJ, McPherson R, Willer CJ, Erdmann J, Hall AS, Samani NJ, Deloukas P, Schunkert H, Wilson JG, Kooperberg C, Rich SS, Tracy RP, Lin DY, Altshuler D, Gabriel S, Nickerson DA, Jarvik GP, Cupples LA, Reiner AP, Boerwinkle E, Kathiresan S. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med 2014;371:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K.. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491–1493. [DOI] [PubMed] [Google Scholar]
- 19. McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC.. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann H-E, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet D-A, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H.. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O'Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ.. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators, Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, König IR, Weeke PE, Webb TR, Auer PL, Schick UM, Lu Y, Zhang H, Dube MP, Goel A, Farrall M, Peloso GM, Won HH, Do R, van Iperen E, Kanoni S, Kruppa J, Mahajan A, Scott RA, Willenberg C, Braund PS, van Capelleveen JC, Doney AS, Donnelly LA, Asselta R, Merlini PA, Duga S, Marziliano N, Denny JC, Shaffer CM, El-Mokhtari NE, Franke A, Gottesman O, Heilmann S, Hengstenberg C, Hoffman P, Holmen OL, Hveem K, Jansson JH, Jöckel KH, Kessler T, Kriebel J, Laugwitz KL, Marouli E, Martinelli N, McCarthy MI, Van Zuydam NR, Meisinger C, Esko T, Mihailov E, Escher SA, Alver M, Moebus S, Morris AD, Müller-Nurasyid M, Nikpay M, Olivieri O, Lemieux Perreault LP, AlQarawi A, Robertson NR, Akinsanya KO, Reilly DF, Vogt TF, Yin W, Asselbergs FW, Kooperberg C, Jackson RD, Stahl E, Strauch K, Varga TV, Waldenberger M, Zeng L, Kraja AT, Liu C, Ehret GB, Newton-Cheh C, Chasman DI, Chowdhury R, Ferrario M, Ford I, Jukema JW, Kee F, Kuulasmaa K, Nordestgaard BG, Perola M, Saleheen D, Sattar N, Surendran P, Tregouet D, Young R, Howson JM, Butterworth AS, Danesh J, Ardissino D, Bottinger EP, Erbel R, Franks PW, Girelli D, Hall AS, Hovingh GK, Kastrati A, Lieb W, Meitinger T, Kraus WE, Shah SH, McPherson R, Orho-Melander M, Melander O, Metspalu A, Palmer CN, Peters A, Rader D, Reilly MP, Loos RJ, Reiner AP, Roden DM, Tardif JC, Thompson JR, Wareham NJ, Watkins H, Willer CJ, Kathiresan S, Deloukas P, Samani NJ, Schunkert H. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N Engl J Med 2016;374:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howson JMM, Zhao W, Barnes DR, Ho W-K, Young R, Paul DS, Waite LL, Freitag DF, Fauman EB, Salfati EL, Sun BB, Eicher JD, Johnson AD, Sheu WHH, Nielsen SF, Lin W-Y, Surendran P, Malarstig A, Wilk JB, Tybjærg-Hansen A, Rasmussen KL, Kamstrup PR, Deloukas P, Erdmann J, Kathiresan S, Samani NJ, Schunkert H, Watkins H, Do R, Rader DJ, Johnson JA, Hazen SL, Quyyumi AA, Spertus JA, Pepine CJ, Franceschini N, Justice A, Reiner AP, Buyske S, Hindorff LA, Carty CL, North KE, Kooperberg C, Boerwinkle E, Young K, Graff M, Peters U, Absher D, Hsiung CA, Lee W-J, Taylor KD, Chen Y-H, Lee I-T, Guo X, Chung R-H, Hung Y-J, Rotter JI, Juang J-MJ, Quertermous T, Wang T-D, Rasheed A, Frossard P, Alam DS, Majumder AAS, Di Angelantonio E, Chowdhury R, Chen Y-DI, Nordestgaard BG, Assimes TL, Danesh J, Butterworth AS, Saleheen D.. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet 2017;49:1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, Segre AV, Ardlie KG, Khera AV, Kaushik VK, Natarajan P, Kathiresan S.. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet 2017;49:1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimaki T, Lu X, Lu Y, Marz W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Bjorkegren JLM, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H, Deloukas P.. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385–1391. [DOI] [PubMed] [Google Scholar]
- 26. Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ.. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verweij N, Eppinga RN, Hagemeijer Y, van der Harst P.. Identification of 15 novel risk loci for coronary artery disease and genetic risk of recurrent events, atrial fibrillation and heart failure. Sci Rep 2017;7:2761.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clarke SL, Assimes TL.. Genome-wide association studies of coronary artery disease: recent progress and challenges ahead. Curr Atheroscler Rep 2018;20:47.. [DOI] [PubMed] [Google Scholar]
- 29. van der Harst P, Verweij N.. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O'Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM.. Genome-wide association study of blood pressure and hypertension. Nat Genet 2009;41:677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O'Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O'Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB.. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 2009;41:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Doring A, Elliott P, Epstein SE, Ingi Eyjolfsson G, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimaki T, Lin SY, Lindstrom J, Loos RJF, Mach F, McArdle WL, Meisinger C, Mitchell BD, Muller G, Nagaraja R, Narisu N, Nieminen TVM, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancakova A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YI, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrieres J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Jarvelin MR, Jula A, Kahonen M, Kaprio J, Kesaniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, Marz W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njolstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PEH, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BHR, Ordovas JM, Boerwinkle E, Palmer CNA, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR.. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen M-R, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn G-W, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S.. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336. [DOI] [PubMed] [Google Scholar]
- 34. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M.. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P.. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885. [DOI] [PubMed] [Google Scholar]
- 36. Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki M-L, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S.. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 2010;376:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tada H, Melander O, Louie JZ, Catanese JJ, Rowland CM, Devlin JJ, Kathiresan S, Shiffman D.. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J 2016;37:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abraham G, Havulinna AS, Bhalala OG, Byars SG, De Livera AM, Yetukuri L, Tikkanen E, Perola M, Schunkert H, Sijbrands EJ, Palotie A, Samani NJ, Salomaa V, Ripatti S, Inouye M.. Genomic prediction of coronary heart disease. Eur Heart J 2016;37:3267–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ.. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010;466:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cervelli T, Borghini A, Galli A, Andreassi MG.. DNA damage and repair in atherosclerosis: current insights and future perspectives. Int J Mol Sci 2012;13:16929–16944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Libby P. Inflammation in atherosclerosis. Nature 2002;420:868–874. [DOI] [PubMed] [Google Scholar]
- 42. Lilly LS, Braunwald E.. Braunwald's Heart Disease: A Textbook of Cardiovascular Medicine. Philadelphia, PA: Elsevier Health Sciences; 2012. [Google Scholar]
- 43. Wang JC, Bennett M.. Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ Res 2012;111:245–259. [DOI] [PubMed] [Google Scholar]
- 44. Martinet W, Knaapen MWM, De Meyer GRY, Herman AG, Kockx MM.. Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation 2002;106:927–932. [DOI] [PubMed] [Google Scholar]
- 45. Ballinger SW, Patterson C, Knight-Lozano CA, Burow DL, Conklin CA, Hu Z, Reuf J, Horaist C, Lebovitz R, Hunter GC, McIntyre K, Runge MS.. Mitochondrial integrity and function in atherogenesis. Circulation 2002;106:544–549. [DOI] [PubMed] [Google Scholar]
- 46. Botto N, Berti S, Manfredi S, Al-Jabri A, Federici C, Clerico A, Ciofini E, Biagini A, Andreassi MG.. Detection of mtDNA with 4977 bp deletion in blood cells and atherosclerotic lesions of patients with coronary artery disease. Mutat Res 2005;570:81–88. [DOI] [PubMed] [Google Scholar]
- 47. Yu E, Calvert PA, Mercer JR, Harrison J, Baker L, Figg NL, Kumar S, Wang JC, Hurst LA, Obaid DR, Logan A, West NE, Clarke MC, Vidal-Puig A, Murphy MP, Bennett MR.. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation 2013;128:702–712. [DOI] [PubMed] [Google Scholar]
- 48. Hatzistamou J, Kiaris H, Ergazaki M, Spandidos D.. Loss of heterozygosity and microsatellite instability in human atherosclerotic plaques. Biochem Biophys Res Commun 1996;225:186–190. [DOI] [PubMed] [Google Scholar]
- 49. Kiaris H, Hatzistamou J, Spandidos DA.. Instability at the H-ras minisatellite in human atherosclerotic plaques. Atherosclerosis 1996;125:47–51. [DOI] [PubMed] [Google Scholar]
- 50. McCaffrey TA, Du B, Consigli S, Szabo P, Bray PJ, Hartner L, Weksler BB, Sanborn TA, Bergman G, Bush HL.. Genomic instability in the type II TGF-beta1 receptor gene in atherosclerotic and restenotic vascular cells. J Clin Invest 1997;100:2182–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Casalone R, Granata P, Minelli E, Portentoso P, Giudici A, Righi R, Castelli P, Socrate A, Frigerio B.. Cytogenetic analysis reveals clonal proliferation of smooth muscle cells in atherosclerotic plaques. Hum Genet 1991;87:139–143. [DOI] [PubMed] [Google Scholar]
- 52. Brouilette SW, Moore JS, McMahon AD, Thompson JR, Ford I, Shepherd J, Packard CJ, Samani NJ.. Telomere length, risk of coronary heart disease, and statin treatment in the West of Scotland Primary Prevention Study: a nested case-control study. Lancet 2007;369:107–114. [DOI] [PubMed] [Google Scholar]
- 53. Salpea KD, Talmud PJ, Cooper JA, Maubaret CG, Stephens JW, Abelak K, Humphries SE.. Association of telomere length with type 2 diabetes, oxidative stress and UCP2 gene variation. Atherosclerosis 2010;209:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Willeit P, Willeit J, BrandstäTter A, Ehrlenbach S, Mayr A, Gasperi A, Weger S, Oberhollenzer F, Reindl M, Kronenberg F, Kiechl S.. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler Thromb Vasc Biol 2010;30:1649–1656. [DOI] [PubMed] [Google Scholar]
- 55. Herbert KE, Mistry Y, Hastings R, Poolman T, Niklason L, Williams B.. Angiotensin II-mediated oxidative DNA damage accelerates cellular senescence in cultured human vascular smooth muscle cells via telomere-dependent and independent pathways. Circ Res 2008;102:201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahmoudi M, Gorenne I, Mercer J, Figg N, Littlewood T, Bennett M.. Statins use a novel nijmegen breakage syndrome-1-dependent pathway to accelerate DNA repair in vascular smooth muscle cells. Circ Res 2008;103:717–725. [DOI] [PubMed] [Google Scholar]
- 57. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L.. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landen M, Hoglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Gronberg H, Hultman CM, McCarroll SA.. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL.. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jan M, Ebert BL, Jaiswal S.. Clonal hematopoiesis. Semin Hematol 2017;54:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Natarajan P, Jaiswal S, Kathiresan S.. Clonal hematopoiesis: somatic mutations in blood cells and atherosclerosis. Circ Genom Precis Med 2018;11:e001926.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL.. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, Jonasson JG, Tryggvadottir L, Jonsson T, Helgason A, Gylfason A, Sulem P, Rafnar T, Thorsteinsdottir U, Gudbjartsson DF, Masson G, Kong A, Stefansson K.. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017;130:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL.. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu C-L, Sano S, Muralidharan S, Rius C, Vuong J, Jacob S, Muralidhar V, Robertson AAB, Cooper MA, Andrés V, Hirschi KK, Martin KA, Walsh K.. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017;355:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner M-J, Cullen M, Epstein CG, Burdett L, Dean MC, Chatterjee N, Sampson J, Chung CC, Kovaks J, Gapstur SM, Stevens VL, Teras LT, Gaudet MM, Albanes D, Weinstein SJ, Virtamo J, Taylor PR, Freedman ND, Abnet CC, Goldstein AM, Hu N, Yu K, Yuan J-M, Liao L, Ding T, Qiao Y-L, Gao Y-T, Koh W-P, Xiang Y-B, Tang Z-Z, Fan J-H, Aldrich MC, Amos C, Blot WJ, Bock CH, Gillanders EM, Harris CC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, McNeill LH, Rybicki BA, Schwartz AG, Signorello LB, Spitz MR, Wiencke JK, Wrensch M, Wu X, Zanetti KA, Ziegler RG, Figueroa JD, Garcia-Closas M, Malats N, Marenne G, Prokunina-Olsson L, Baris D, Schwenn M, Johnson A, Landi MT, Goldin L, Consonni D, Bertazzi PA, Rotunno M, Rajaraman P, Andersson U, Beane Freeman LE, Berg CD, Buring JE, Butler MA, Carreon T, Feychting M, Ahlbom A, Gaziano JM, Giles GG, Hallmans G, Hankinson SE, Hartge P, Henriksson R, Inskip PD, Johansen C, Landgren A, McKean-Cowdin R, Michaud DS, Melin BS, Peters U, Ruder AM, Sesso HD, Severi G, Shu X-O, Visvanathan K, White E, Wolk A, Zeleniuch-Jacquotte A, Zheng W, Silverman DT, Kogevinas M, Gonzalez JR, Villa O, Li D, Duell EJ, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Hassan M, Wheeler W, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross MD, Holly EA, Klein AP, LaCroix A, Mandelson MT, Petersen G, Boutron-Ruault M-C, Bracci PM, Canzian F, Chang K, Cotterchio M, Giovannucci EL, Goggins M, Hoffman Bolton JA, Jenab M, Khaw K-T, Krogh V, Kurtz RC, McWilliams RR, Mendelsohn JB, Rabe KG, Riboli E, Tjønneland A, Tobias GS, Trichopoulos D, Elena JW, Yu H, Amundadottir L, Stolzenberg-Solomon RZ, Kraft P, Schumacher F, Stram D, Savage SA, Mirabello L, Andrulis IL, Wunder JS, Patiño García A, Sierrasesúmaga L, Barkauskas DA, Gorlick RG, Purdue M, Chow W-H, Moore LE, Schwartz KL, Davis FG, Hsing AW, Berndt SI, Black A, Wentzensen N, Brinton LA, Lissowska J, Peplonska B, McGlynn KA, Cook MB, Graubard BI, Kratz CP, Greene MH, Erickson RL, Hunter DJ, Thomas G, Hoover RN, Real FX, Fraumeni JF, Caporaso NE, Tucker M, Rothman N, Pérez-Jurado LA, Chanock SJ.. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012;44:651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Loh PR, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, Birmann BM, Talkowski ME, Bakhoum SF, McCarroll SA, Price AL.. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018;559:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bonnefond A, Skrobek B, Lobbens S, Eury E, Thuillier D, Cauchi S, Lantieri O, Balkau B, Riboli E, Marre M, Charpentier G, Yengo L, Froguel P.. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat Genet 2013;45:1040–1043. [DOI] [PubMed] [Google Scholar]
- 69.Institute NHGR. The Cost of Sequencing a Human Genome National Human Genome Research Institute, July 6, 2016. https://www.genome.gov/27565109/the-cost-of-sequencing-a-human-genome/ (12 February 2019, date last accessed).
- 70. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X.. The sequence of the human genome. Science 2001;291:1304–1351. [DOI] [PubMed] [Google Scholar]
- 71. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J.. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 72. Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard DS, Guttmacher A, Guyer M, Hemsley FM, Jennings JL, Kerr D, Klatt P, Kolar P, Kusada J, Lane DP, Laplace F, Youyong L, Nettekoven G, Ozenberger B, Peterson J, Rao TS, Remacle J, Schafer AJ, Shibata T, Stratton MR, Vockley JG, Watanabe K, Yang H, Yuen MMF, Knoppers BM, Bobrow M, Cambon-Thomsen A, Dressler LG, Dyke SOM, Joly Y, Kato K, Kennedy KL, Nicolás P, Parker MJ, Rial-Sebbag E, Romeo-Casabona CM, Shaw KM, Wallace S, Wiesner GL, Zeps N, Lichter P, Biankin AV, Chabannon C, Chin L, Clément B, de Alava E, Degos F, Ferguson ML, Geary P, Hayes DN, Hudson TJ, Johns AL, Kasprzyk A, Nakagawa H, Penny R, Piris MA, Sarin R, Scarpa A, Shibata T, van de Vijver M, Futreal PA, Aburatani H, Bayés M, Botwell DDL, Campbell PJ, Estivill X, Gerhard DS, Grimmond SM, Gut I, Hirst M, López-Otín C, Majumder P, Marra M, McPherson JD, Nakagawa H, Ning Z, Puente XS, Ruan Y, Shibata T, Stratton MR, Stunnenberg HG, Swerdlow H, Velculescu VE, Wilson RK, Xue HH, Yang L, Spellman PT, Bader GD, Boutros PC, Campbell PJ, Flicek P, Getz G, Guigó R, Guo G, Haussler D, Heath S, Hubbard TJ, Jiang T, Jones SM, Li Q, López-Bigas N, Luo R, Muthuswamy L, Ouellette BFF, Pearson JV, Puente XS, Quesada V, Raphael BJ, Sander C, Shibata T, Speed TP, Stein LD, Stuart JM, Teague JW, Totoki Y, Tsunoda T, Valencia A, Wheeler DA, Wu H, Zhao S, Zhou G, Stein LD, Guigó R, Hubbard TJ, Joly Y, Jones SM, Kasprzyk A, Lathrop M, López-Bigas N, Ouellette BFF, Spellman PT, Teague JW, Thomas G, Valencia A, Yoshida T, Kennedy KL, Axton M, Dyke SOM, Futreal PA, Gerhard DS, Gunter C, Guyer M, Hudson TJ, McPherson JD, Miller LJ, Ozenberger B, Shaw KM, Kasprzyk A, Stein LD, Zhang J, Haider SA, Wang J, Yung CK, Cros A, Cross A, Liang Y, Gnaneshan S, Guberman J, Hsu J, Bobrow M, Chalmers DRC, Hasel KW, Joly Y, Kaan TSH, Kennedy KL, Knoppers BM, Lowrance WW, Masui T, Nicolás P, Rial-Sebbag E, Rodriguez LL, Vergely C, Yoshida T, Grimmond SM, Biankin AV, Bowtell DDL, Cloonan N, deFazio A, Eshleman JR, Etemadmoghadam D, Gardiner BB, Gardiner BA, Kench JG, Scarpa A, Sutherland RL, Tempero MA, Waddell NJ, Wilson PJ, McPherson JD, Gallinger S, Tsao M-S, Shaw PA, Petersen GM, Mukhopadhyay D, Chin L, DePinho RA, Thayer S, Muthuswamy L, Shazand K, Beck T, Sam M, Timms L, Ballin V, Lu Y, Ji J, Zhang X, Chen F, Hu X, Zhou G, Yang Q, Tian G, Zhang L, Xing X, Li X, Zhu Z, Yu Y, Yu J, Yang H, Lathrop M, Tost J, Brennan P, Holcatova I, Zaridze D, Brazma A, Egevard L, Prokhortchouk E, Banks RE, Uhlén M, Cambon-Thomsen A, Viksna J, Ponten F, Skryabin K, Stratton MR, Futreal PA, Birney E, Borg A, Børresen-Dale A-L, Caldas C, Foekens JA, Martin S, Reis-Filho JS, Richardson AL, Sotiriou C, Stunnenberg HG, Thoms G, van de Vijver M, van't Veer L, Calvo F, Birnbaum D, Blanche H, Boucher P, Boyault S, Chabannon C, Gut I, Masson-Jacquemier JD, Lathrop M, Pauporté I, Pivot X, Vincent-Salomon A, Tabone E, Theillet C, Thomas G, Tost J, Treilleux I, Calvo F, Bioulac-Sage P, Clément B, Decaens T, Degos F, Franco D, Gut I, Gut M, Heath S, Lathrop M, Samuel D, Thomas G, Zucman-Rossi J, Lichter P, Eils R, Brors B, Korbel JO, Korshunov A, Landgraf P, Lehrach H, Pfister S, Radlwimmer B, Reifenberger G, Taylor MD, von Kalle C, Majumder PP, Sarin R, Rao TS, Bhan MK, Scarpa A, Pederzoli P, Lawlor RA, Delledonne M, Bardelli A, Biankin AV, Grimmond SM, Gress T, Klimstra D, Zamboni G, Shibata T, Nakamura Y, Nakagawa H, Kusada J, Tsunoda T, Miyano S, Aburatani H, Kato K, Fujimoto A, Yoshida T, Campo E, López-Otín C, Estivill X, Guigó R, de Sanjosé S, Piris MA, Montserrat E, González-Díaz M, Puente XS, Jares P, Valencia A, Himmelbauer H, Himmelbaue H, Quesada V, Bea S, Stratton MR, Futreal PA, Campbell PJ, Vincent-Salomon A, Richardson AL, Reis-Filho JS, van de Vijver M, Thomas G, Masson-Jacquemier JD, Aparicio S, Borg A, Børresen-Dale A-L, Caldas C, Foekens JA, Stunnenberg HG, van't Veer L, Easton DF, Spellman PT, Martin S, Barker AD, Chin L, Collins FS, Compton CC, Ferguson ML, Gerhard DS, Getz G, Gunter C, Guttmacher A, Guyer M, Hayes DN, Lander ES, Ozenberger B, Penny R, Peterson J, Sander C, Shaw KM, Speed TP, Spellman PT, Vockley JG, Wheeler DA, Wilson RK, Hudson TJ, Chin L, Knoppers BM, Lander ES, Lichter P, Stein LD, Stratton MR, Anderson W, Barker AD, Bell C, Bobrow M, Burke W, Collins FS, Compton CC, DePinho RA, Easton DF, Futreal PA, Gerhard DS, Green AR, Guyer M, Hamilton SR, Hubbard TJ, Kallioniemi OP, Kennedy KL, Ley TJ, Liu ET, Lu Y, Majumder P, Marra M, Ozenberger B, Peterson J, Schafer AJ, Spellman PT, Stunnenberg HG, Wainwright BJ, Wilson RK, Yang H.. International network of cancer genome projects. Nature 2010;464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, Guarino P, Aslan M, Anderson D, LaFleur R, Hammond T, Schaa K, Moser J, Huang G, Muralidhar S, Przygodzki R, O'Leary TJ.. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016;70:214–223. [DOI] [PubMed] [Google Scholar]
- 74. Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR.. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gainer V, Cagan A, Castro V, Duey S, Ghosh B, Goodson A, Goryachev S, Metta R, Wang T, Wattanasin N, Murphy S.. The Biobank Portal for Partners personalized medicine: a query tool for working with consented biobank samples, genotypes, and phenotypes using i2b2. J Pers Med 2016;6:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R.. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patel AP, Peloso GM, Pirruccello JP, Johansen CT, Dubé JB, Larach DB, Ban MR, Dallinge-Thie GM, Gupta N, Boehnke M, Abecasis GR, Kastelein JJP, Hovingh GK, Hegele RA, Rader DJ, Kathiresan S.. Targeted exonic sequencing of GWAS loci in the high extremes of the plasma lipids distribution. Atherosclerosis 2016;250:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Consortium E. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S.. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Visscher H, Ross CJ, Rassekh SR, Barhdadi A, Dube MP, Al-Saloos H, Sandor GS, Caron HN, van Dalen EC, Kremer LC, van der Pal HJ, Brown AM, Rogers PC, Phillips MS, Rieder MJ, Carleton BC, Hayden MR.. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol 2012;30:1422–1428. [DOI] [PubMed] [Google Scholar]
- 81. Ky B, Putt M, Sawaya H, French B, Januzzi JL Jr, Sebag IA, Plana JC, Cohen V, Banchs J, Carver JR, Wiegers SE, Martin RP, Picard MH, Gerszten RE, Halpern EF, Passeri J, Kuter I, Scherrer-Crosbie M.. Early increases in multiple biomarkers predict subsequent cardiotoxicity in patients with breast cancer treated with doxorubicin, taxanes, and trastuzumab. J Am Coll Cardiol 2014;63:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, Sartori S, Fuster V, Reilly DF, Butterworth A, Rader DJ, Ford I, Sattar N, Kathiresan S.. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 2017;135:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yizhak K, Aguet F, Kim J, Hess J, Kubler K, Grimsby J, Frazer R, Zhang H, Haradhvala N, Rosebrock D, Livitz D, Li X, Arich-Landkof E, Shoresh N, Stewart C, Segre A, Branton P, Polak P, Ardlie K, Getz G.. A comprehensive analysis of RNA sequences reveals macroscopic somatic clonal expansion across normal tissues. bioRxiv 2018:416339. https://www.biorxiv.org/content/10.1101/416339v1.abstract (12 February 2019, date last accessed). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. van den Belt-Dusebout AW, Nuver J, de Wit R, Gietema JA, ten Bokkel Huinink WW, Rodrigus PT, Schimmel EC, Aleman BM, van Leeuwen FE.. Long-term risk of cardiovascular disease in 5-year survivors of testicular cancer. J Clin Oncol 2006;24:467–475. [DOI] [PubMed] [Google Scholar]
- 85. Haugnes HS, Wethal T, Aass N, Dahl O, Klepp O, Langberg CW, Wilsgaard T, Bremnes RM, Fossa SD.. Cardiovascular risk factors and morbidity in long-term survivors of testicular cancer: a 20-year follow-up study. J Clin Oncol 2010;28:4649–4657. [DOI] [PubMed] [Google Scholar]
- 86. Alter P, Herzum M, Soufi M, Schaefer JR, Maisch B.. Cardiotoxicity of 5-fluorouracil. Cardiovasc Hematol Agents Med Chem 2006;4:1–5. [DOI] [PubMed] [Google Scholar]
- 87. Scappaticci FA, Skillings JR, Holden SN, Gerber HP, Miller K, Kabbinavar F, Bergsland E, Ngai J, Holmgren E, Wang J, Hurwitz H.. Arterial thromboembolic events in patients with metastatic carcinoma treated with chemotherapy and bevacizumab. J Natl Cancer Inst 2007;99:1232–1239. [DOI] [PubMed] [Google Scholar]
- 88. Lenneman CG, Sawyer DB.. Cardio-oncology: an update on cardiotoxicity of cancer-related treatment. Circ Res 2016;118:1008–1020. [DOI] [PubMed] [Google Scholar]
- 89. Basavaraju SR, Easterly CE.. Pathophysiological effects of radiation on atherosclerosis development and progression, and the incidence of cardiovascular complications. Med Phys 2002;29:2391–2403. [DOI] [PubMed] [Google Scholar]
- 90. Okano M, Bell DW, Haber DA, Li E.. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257. [DOI] [PubMed] [Google Scholar]
- 91. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O'Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK.. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010;363:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR, Committee FS.. Investigators Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 2017;376:1713–1722. [DOI] [PubMed] [Google Scholar]
- 93. Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, Edelberg JM, Goodman SG, Hanotin C, Harrington RA, Jukema JW, Lecorps G, Mahaffey KW, Moryusef A, Pordy R, Quintero K, Roe MT, Sasiela WJ, Tamby JF, Tricoci P, White HD, Zeiher AM.. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med 2018;379:2097–2107. [DOI] [PubMed] [Google Scholar]
- 94. Dewey FE, Gusarova V, Dunbar RL, O'Dushlaine C, Schurmann C, Gottesman O, McCarthy S, Van Hout CV, Bruse S, Dansky HM, Leader JB, Murray MF, Ritchie MD, Kirchner HL, Habegger L, Lopez A, Penn J, Zhao A, Shao W, Stahl N, Murphy AJ, Hamon S, Bouzelmat A, Zhang R, Shumel B, Pordy R, Gipe D, Herman GA, Sheu WHH, Lee I-T, Liang K-W, Guo X, Rotter JI, Chen Y-DI, Kraus WE, Shah SH, Damrauer S, Small A, Rader DJ, Wulff AB, Nordestgaard BG, Tybjærg-Hansen A, van den Hoek AM, Princen HMG, Ledbetter DH, Carey DJ, Overton JD, Reid JG, Sasiela WJ, Banerjee P, Shuldiner AR, Borecki IB, Teslovich TM, Yancopoulos GD, Mellis SJ, Gromada J, Baras A.. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N Engl J Med 2017;377:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Graham MJ, Lee RG, Brandt TA, Tai LJ, Fu W, Peralta R, Yu R, Hurh E, Paz E, McEvoy BW, Baker BF, Pham NC, Digenio A, Hughes SG, Geary RS, Witztum JL, Crooke RM, Tsimikas S.. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med 2017;377:222–232. [DOI] [PubMed] [Google Scholar]
- 96. Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, Braun LTD, Ferranti S, Faiella-Tommasino J, Forman DE, Goldberg R, Heidenreich PA, Hlatky MA, Jones DW, Lloyd-Jones D, Lopez-Pajares N, Ndumele CE, Orringer CE, Peralta CA, Saseen JJ, Smith SC Jr, Sperling L, Virani SS, Yeboah J.. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the management of blood cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2018;25709. Doi:10.1016/j.jacc.2018.11.003. [Google Scholar]
- 97. Rath MG, Uhlmann L, Fiedler M, Heil J, Golatta M, Dinkic C, Hennigs A, Schott S, Ernst V, Koch T, Sohn C, Brucker C, Rom J.. Oncotype DX® in breast cancer patients: clinical experience, outcome and follow-up-a case-control study. Arch Gynecol Obstet 2018;297:443–447. [DOI] [PubMed] [Google Scholar]
- 98. Heist RS, Engelman JA.. SnapShot: non-small cell lung cancer. Cancer Cell 2012;21:448.e2. [DOI] [PubMed] [Google Scholar]
- 99. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A.. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472–484. [DOI] [PubMed] [Google Scholar]
- 100. Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, Grillo I, Bakogianni S, Ndiaye-Lobry D, Martín MT, Guillamot M, Banh RS, Xu M, Figueroa ME, Dickins RA, Abdel-Wahab O, Park CY, Tsirigos A, Neel BG, Aifantis I.. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 2017;170:1079–1095.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ridker PM, Thuren T, Zalewski A, Libby P.. Interleukin-1β inhibition and the prevention of recurrent cardiovascular events: rationale and design of the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS). Am Heart J 2011;162:597–605. [DOI] [PubMed] [Google Scholar]
- 102. Svensson EC, Madar A, Campbell CD, He Y, Sultan M, Healey ML, D’Aco K, Fernandez A, Wache-Mainier C, Ridker PM.. TET2-driven clonal hematopoiesis predicts enhanced response to canakinumab in the CANTOS trial: an exploratory analysis. Circulation 2018;138:A15111–A15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Musunuru K. How genome editing could be used in the treatment of cardiovascular diseases. Per Med 2018;15:67–69. [DOI] [PubMed] [Google Scholar]