FIGURE 1.
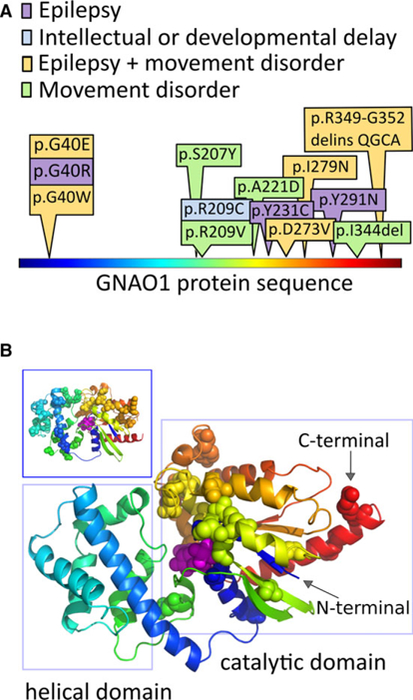
Patient variants in GNAO1 affect the guanosine triphosphate–binding region. Disease‐associated amino acid residues in the GNAO1 protein localize to the catalytic domain. A, The protein is shown in linear representation, colored using a “rainbow” scheme, starting with blue at the N‐terminal, and ending with red at the C‐terminal. The amino acid substitutions altered by the GNAO1 variants in our series are depicted here, with colors depicting the neurological features present in each case. B, The positions of disease‐associated variants, from this series and the literature, are shown as spheres on the model of the GNAO1 structure. The GNAO1 substrate is also shown as spheres, colored magenta. The same coloring scheme is used in A and B. Patient variants cluster in the region of the catalytic domain (right rectangle), which starts at the very N‐terminal aspect of the sequence, weaves into the helical domain, and returns back to complete the catalytic domain structure. The long helix on the N‐terminal is not shown, because its position in the active conformation of Gα is uncertain, and it carries no reported disease‐associated amino acid positions. Inset (upper left-hand box): The distribution of missense variant amino acids in ExAC17 covers both structural domains of the protein and does not overlap with the disease‐associated amino acid changes in our series
