Fig. 3.
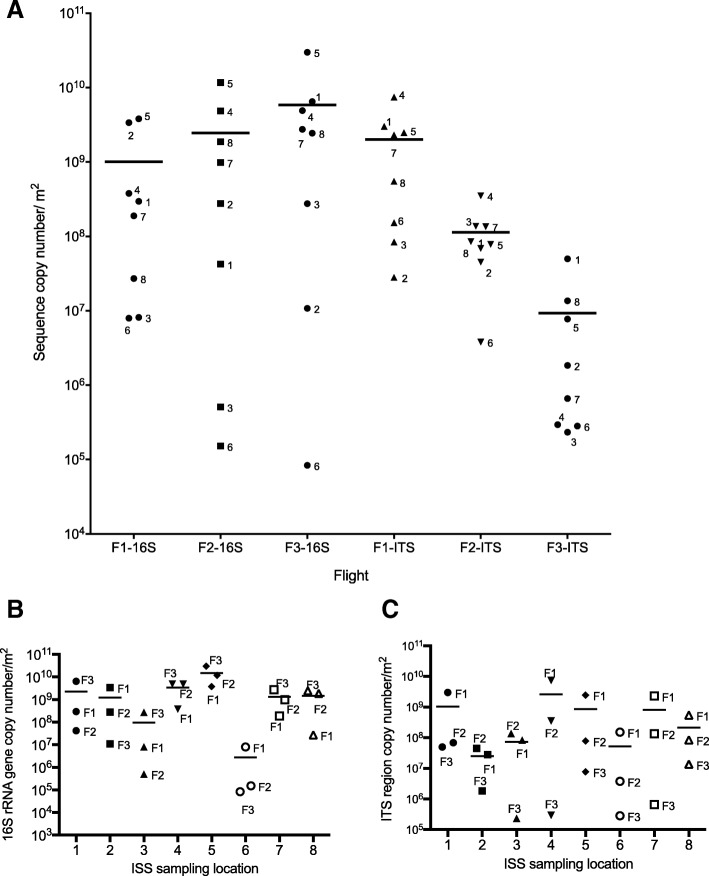
Intact cell membrane/viable bacterial and fungal population aboard the ISS as estimated by PMA-qPCR. a Scatter plot comparing the 16S rRNA gene (bacteria) and ITS region (fungi) copy numbers of PMA treated samples collected during flights 1, 2, and 3. Each column represents a single flight and each symbol in a column (labeled with a number) represents one of the eight locations sampled during that flight. The horizontal line in each column represents the average gene copy number/m2 for each Flight. b Scatter plot comparing 16S rRNA gene and c ITS region (fungi) copy numbers across locations. Each column “L” followed by a number represents a location and each dot in a column represents the flight it was sampled from. The horizontal line in each column represents the average copy number/m2 at that location. NB: The 16S rRNA gene copy number was not adjusted to the average number per bacterial genome. Control samples were measured and found to be at the level of 102 16S rRNA gene copies per μL. Even when the initial template volume was increased to 10 μL, the expected 20-fold increase in the gene copy numbers was not observed. In panel a, F1-ITS was statistically significantly higher than F3-ITS (P < 0.05). No statistically significant differences were observed in panel b (P > 0.05). The statistical test was performed with the Kruskal-Wallis test followed by Dunn’s post-hoc test