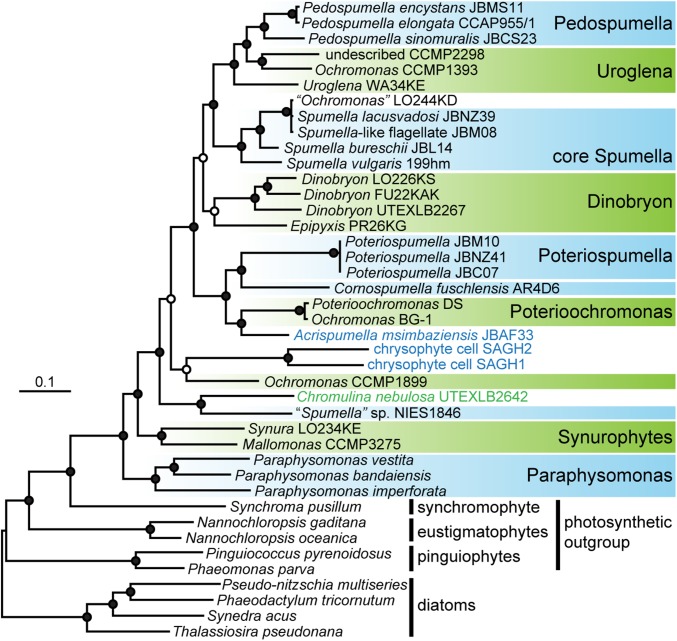
Fig. 1.
Multigene Bayesian consensus phylogeny of chrysophytes inferred from 17,439 amino acid sites under three substitution models. Photosynthetic and secondarily nonphotosynthetic chrysophyte lineages are indicated in green and blue, respectively; the trophic status of Ochromonas LO244KD remains debated (15, 21). Taxa used for subsequent plastid metabolism comparisons are contained in boxes. Filled circles at nodes indicate Bayesian posterior probabilities of 1.0 and maximum likelihood bootstrap support greater than 80% for all analyses. Open circles at nodes indicate Bayesian posterior probabilities greater than 0.8 for all analyses. The topology is displayed following (5), with the root between the diatoms and the PESC clade (pinguiophytes, eustigmatophytes, synchromophytes, and chrysophytes). A corresponding 18S rDNA tree is shown in SI Appendix, Fig. S1.