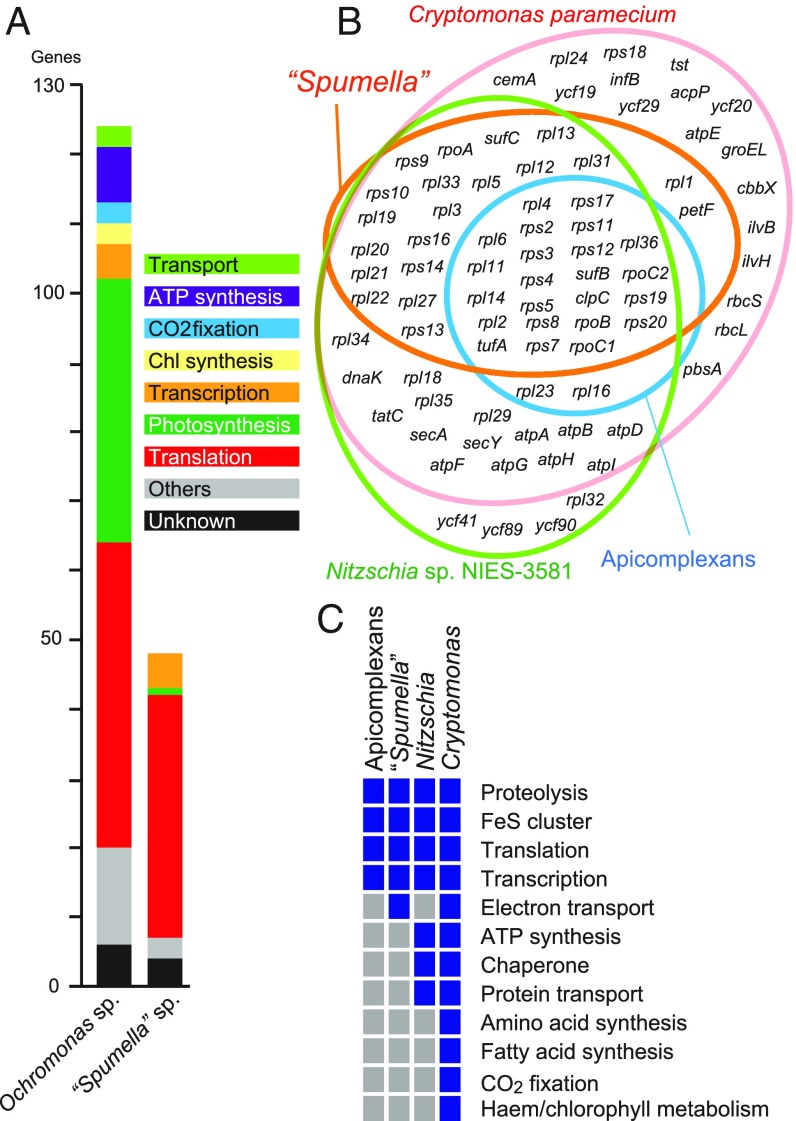
Fig. 2.
Plastid genome of “Spumella” sp. NIES-1846. (A) Comparison of protein-coding genes between a photosynthetic chrysophyte and “Spumella” sp. NIES-1846 (22). Each color bar shows a functional category. (B) Venn diagram of protein-coding genome contents in various nonphotosynthetic, red-alga–derived plastid lineages. Nitzschia sp. NIES-3581 (3) and Cryptomonas paramecium (4) are used as representatives for nonphotosynthetic diatoms and cryptomonads. (C) Plastid-encoded functions in four nonphotosynthetic, red-alga–derived plastid lineages. Blue and gray boxes indicate presence and absence, respectively. A complete plastid genome map is provided in SI Appendix, Fig. S2; comparisons of plastid coding content in a wider range of nonphotosynthetic species are shown in SI Appendix, Fig. S3; and schematic reconstructions and exemplar localizations of plastid metabolism pathways are in SI Appendix, Figs. S4–S6.