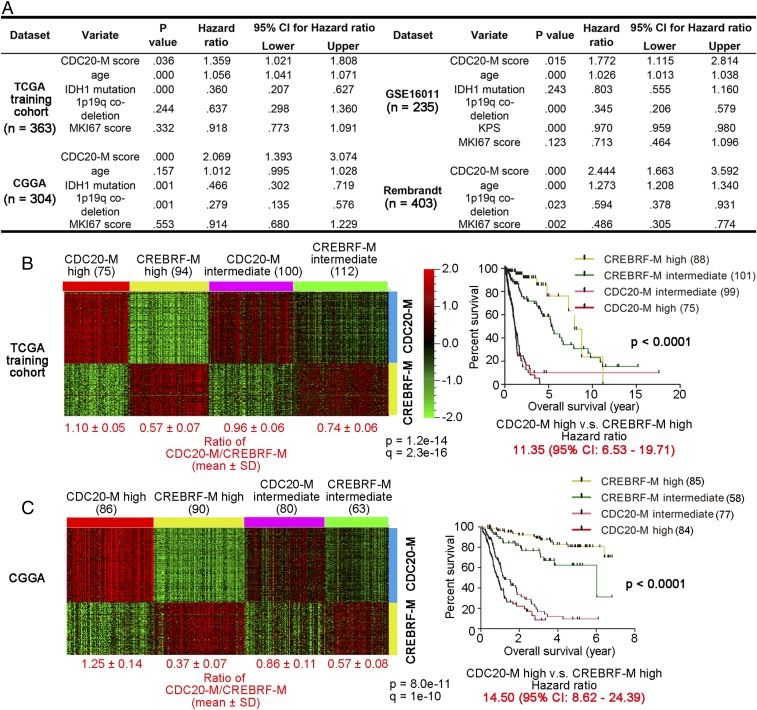
Fig. 2.
Elevated CDC20-M expression is a superior and robust signature of poor prognosis. (A) Results of multivariate Cox regression analysis for CDC20-M score, age at diagnosis, IDH1 mutation, 1p19q codeletion, and MKI67 score (the log2 RNA expression of Ki-67) in four large glioma datasets. Only the CDC20-M score showed consistent correlation with patient survival. (B, Left) Heatmap of unsupervised consensus clustering of the CDC20-M and CREBRF-M signature in the TCGA training cohort. The ratios of average expression of CDC20-M and CREBRF-M are shown under each subgroup. (Right) Kaplan–Meier plot of the overall survival for patients from each molecular subtype is shown. The overall survival data were analyzed using log-rank tests. The hazard ratio between patients with high CDC20-M glioma and patients with high CREBRF-M gliomas is shown under the survival curves. (C, Left) Heatmap of the CDC20-M and CREBRF-M signature in the CGGA glioma dataset supervised by SSP prediction of the CDC20-M/CREBRF-M subtypes. Using Spearman correlation, CGGA glioma samples were assigned to the nearest centroid of the four CDC20-M/CREBRF-M subtypes defined in the TCGA training cohort. The ratios of average expression of CDC20-M and CREBRF-M are shown under each subgroup. (C, Right) Kaplan–Meier plot of the overall survival for patients from each molecular subtype is shown. The overall survival data were analyzed using log-rank tests. The hazard ratio between patients with high CDC20-M glioma and patients with high CREBRF-M gliomas is shown under the survival curves.