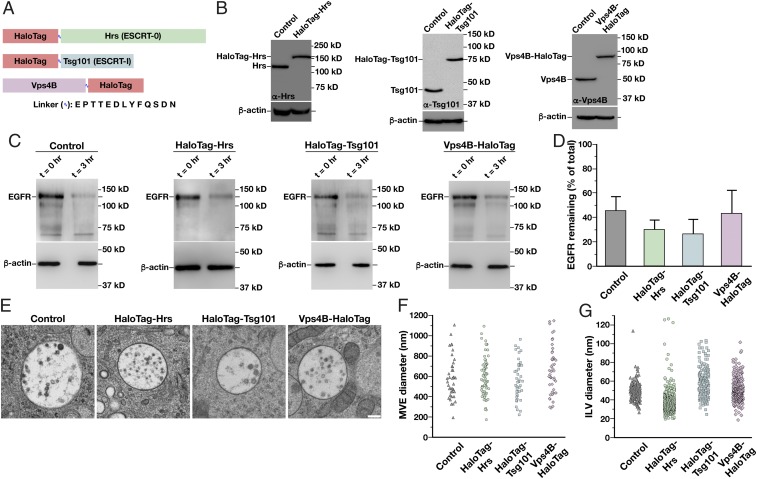
Fig. 1.
Generation of functionally tagged ESCRT subunits using CRISPR/Cas9-mediated genome editing. (A) Cartoon highlighting the placement of the HaloTag on three ESCRT subunits to track ESCRT-0 (green), ESCRT-I (blue), and Vps4 (purple) dynamics. The sequence of the flexible linker used in each case, which contains the tobacco etch virus protease recognition sequence, is also shown. (B) Representative immunoblot analyses of control and CRISPR/Cas9-modified cell lines (n = 3 each) using antibodies directed against Hrs (Left), Tsg101 (Center), Vps4B (Right), and β-actin (load control). (C) Control and CRISPR/Cas9-modified clonal cell lines were incubated in the presence or absence of 30 ng/mL EGF for 3 h following serum starvation, and extracts were immunoblotted using antibodies directed against EGFR (Top) and β-actin (Bottom, load control). (D) Quantification of the percentage of EGFR remaining after 3 h of EGF treatment in control and CRISPR/Cas9-modified clonal cell lines. Error bars represent mean ± SEM (n = 4 each). No statistically significant difference was found, as calculated using an ANOVA test. (E) Representative thin-section EM images of MVEs from control and CRISPR/Cas9-modified clonal cell lines (more than 35 MVEs and 200 ILVs examined in each). (Scale bar: 200 nm.) The size distribution of MVEs (F) and ILVs (G) in control and CRISPR/Cas9-modified clonal cell lines is shown. No statistically significant differences were found, as calculated using an ANOVA test.