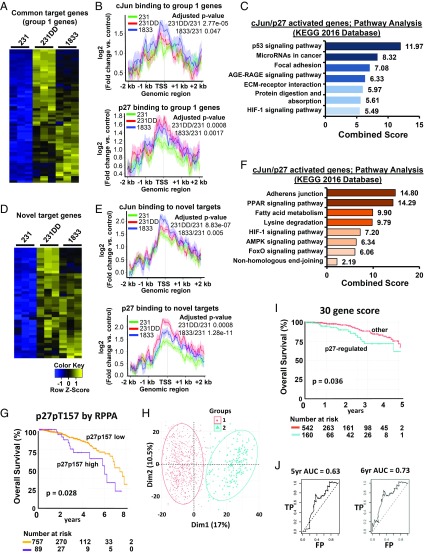
Fig. 7.
cJun/p27-regulated target genes govern cancer programs of tumor progression. (A) Heatmap of expression of 61 p27/cJun target genes common to all three lines whose expression is up-regulated in 231p27CK−DD (231DD) and 1833 lines vs. 231. See also SI Appendix, Table S4. (B) Mean amplitudes of target gene binding by cJun (Top) and p27 (Bottom) are shown for the differentially expressed genes in A in the indicated cell lines. (C) GO analysis shows the top signaling pathways related to the differentially expressed genes identified in A. (D) Heatmap of expression of 43 newly acquired cJun/p27 target genes bound in 231DD and 1833, but not in 231, that are differentially expressed in the indicated cell lines. See also SI Appendix, Table S5. (E) Mean amplitude of target gene binding by cJun (Top) and p27 (Bottom) are shown for the differentially expressed genes in D in the indicated cell lines. (F) GO analysis shows the main signaling pathways related to the genes identified in D. See SI Appendix, Fig. S6 for validation of cJun/p27 cotarget genes. (G) Kaplan–Meier (KM) graph showing differential OS among women whose breast cancers have high vs. low p27pT157 on reverse phase protein array (RPPA) from n = 846 cases in the TCGA/TCPA dataset. (H) Principle component analysis of 30 p27-regulated genes expressed among n = 703 breast cancers. (I) KM graph shows overall breast cancer survival according to differential expression of the p27-regulated 30 gene profile among the training set (n = 703 cases). (J) The prognostic value of this p27-regulated 30 gene signature for OS was validated using ROC analysis in an independent breast cancer validation cohort. AGE-RAGE, advanced glycation end products-receptor for advanced glycation end products; AMPK, 5′ AMP-activated protein kinase; FP, false positive; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR, peroxisome proliferator-activated receptor; TP, true positive.