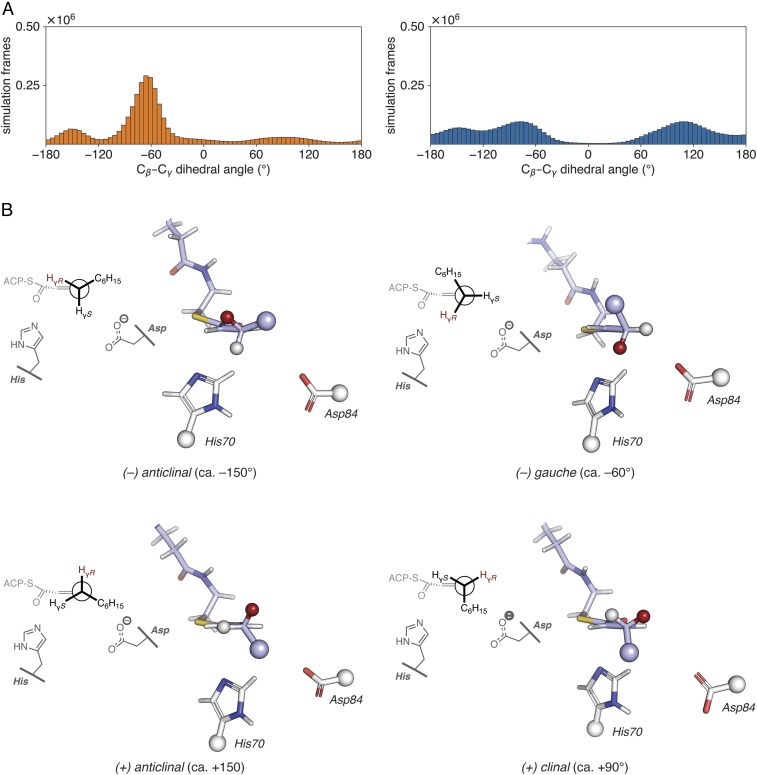
Fig. 6.
Analysis of MD simulations of trans–2-decenoyl-AcpP•DH complexes. (A) Distribution of dihedral angles for the Cɑ–Cβ–Cγ–Cδ torsion of the trans-2-decenoyl substrate sampled throughout the course of GaMD simulations of trans-2-decenoyl-AcpP•FabA (Left, in orange) and trans-dec-2-enoyl-AcpP•FabZ (Right, in blue). Simulation data written every 0.5 ps were binned to prepare histograms using a bin width of 5°. (B) Representative structures of the four Cβ–Cγ rotamers sampled computationally. These structures were identified from the larger simulated ensemble of the trans-2-decenoyl-AcpP•FabA complex as described in SI Appendix. Structures are oriented as Newman projections along the Cγ–Cβ bond of the trans-2-decenoyl substrate (shown in stick representation with carbon, oxygen, and nitrogen atoms in violet, red, and blue, respectively) within the active site of FabA. Catalytic histidine and aspartate are also shown in stick form with white carbon, red oxygen, and blue nitrogen. Large spheres indicate the portions of the substrate and catalytic residues omitted in the 3D images. The smaller spheres found at Cγ show the proR (red) and proS (white) γ-hydrogens; the proR Hγ is abstracted in the isomerization reaction. A 2D representation of each structure is shown on the Left of each panel.