Fig. 3.
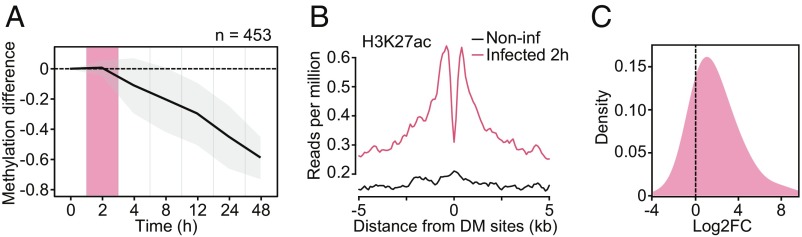
(A) Mean differences in methylation (y-axis) in CpG sites that show stable losses of methylation (similar to Cluster 3 DM sites in Fig. 1 C and D; n = 453) in Salmonella-infected macrophages, across six time points after infection (2, 4, 8, 12, 24, and 48 h [h]; x-axis). Shading denotes ±1 SD. For visualization purposes, we also show the 0 h time point, where we expect no changes in methylation. (B) Composite plots of patterns of H3K27ac ChIP-seq signals ±5 kb around the midpoints of hypomethylated sites (x-axis) in macrophages at 2 h postinfection with Salmonella. (C) Distribution of logtwofold expression changes (between noninfected and Salmonella-infected macrophages at 2 h) for genes associated with DM sites in A (n = 269).