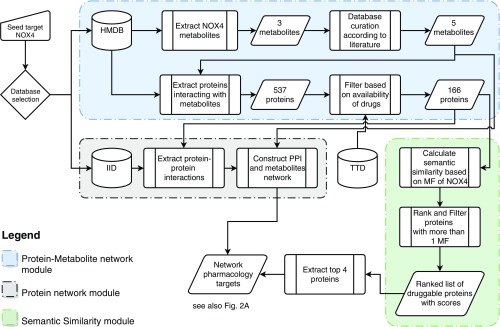
Fig. 1.
Computational workflow for target prioritization via network pharmacology. The computational target prioritization pipeline consists of three interdependent modules. The blue module extracts the metabolites interacting with the protein NOX4 from the Human Metabolome Database, performs curation of the metabolites, extracts the proteins interacting with them, and filters them based on the availability of drugs from the Therapeutic Target Database (TTD). The gray module uses the Integrated Interaction Database (IID) to extract protein–protein interactions of the proteins yielded by the blue module and constructs a network out of them. The green module calculates gene ontology-based semantic similarity scores of the output of the blue module compared with NOX4 using molecular function (MF) annotations, ranks the proteins based on their similarity scores, and excludes proteins with less than one molecular function. The output of the green module is used to annotate the network with the top four proteins.