Abstract
The incidence and geographic range of tick-borne illness associated with Ixodes scapularis and Ixodes pacificus have dramatically increased in recent decades. Anaplasmosis, babesiosis, and Borrelia spirochete infections, including Lyme borreliosis, account for tens of thousands of reported cases of tick-borne disease every year. Assays that reliably detect pathogens in ticks allow investigators and public health agencies to estimate the geographic distribution of human pathogens, assess geographic variation in their prevalence, and evaluate the effectiveness of prevention strategies. As investigators continue to describe new species within the Borrelia burgdorferi sensu lato complex and to recognize some Ixodes-borne Borrelia species as human pathogens, assays are needed to detect and differentiate these species. Here we describe an algorithm to detect and differentiate pathogens in unfed I. scapularis and I. pacificus nymphs including Anaplasma phagocytophilum, Babesia microti, Borrelia burgdorferi sensu stricto, Borrelia mayonii, and Borrelia miyamotoi. The algorithm comprises 5 TaqMan real-time polymerase chain reaction assays and 3 sequencing protocols. It employs multiple targets for each pathogen to optimize specificity, a gene target for I. scapularis and I. pacificus to verify tick-derived DNA quality, and a pan-Borrelia target to detect Borrelia species that may emerge as human disease agents in the future. We assess the algorithm’s sensitivity, specificity, and performance on field-collected ticks.
Keywords: Borrelia, Anaplasma phagocytophilum, Babesia microti, Real-time PCR, Ixodes
1. Introduction
The incidence and geographic range of human tick-borne illnesses caused by pathogens transmitted by the blacklegged tick, Ixodes scapularis, in the eastern United States have dramatically increased in recent decades. The western blacklegged tick, Ixodes pacificus, transmits several of the same pathogens in the far western United States (Eisen et al., 2017). Moreover, new human pathogens associated with these and other tick species continue to emerge in the United States (Eisen et al., 2016, 2017). Ixodes scapularis is an experimentally-confirmed vector of, and is naturally infected with, at least 7 human pathogens: Borrelia burgdorferi sensu stricto (s.s.) and Borrelia mayonii (Lyme borreliosis spirochetes), Borrelia miyamotoi (agent of Borrelia miyamotoi disease, a relapsing fever-like illness), Anaplasma phagocytophilum (agent of anaplasmosis), Babesia microti (agent of babesiosis), Ehrlichia muris subsp. eauclarensis (agent of ehrlichiosis), and Powassan virus lineage 2 (formerly termed deer tick virus, agent of Powassan virus disease) (Bakken and Dumler, 2008; Dolan et al., 2016, 1998; Ebel, 2010; Eisen et al., 2016; Johnson et al., 2015; Karpathy et al., 2016; Krause et al., 2015; Merten and Durden, 2000; Pritt et al., 2017, 2016a, 2016b, 2011; Scoles et al., 2001; Spielman, 1976; Steere et al., 1983; Teglas and Foley, 2006; Vannier and Krause, 2012). Ixodes pacificus is a vector of a subset of the human pathogens that I. scapularis transmits, including B. burgdorferi s.s, A. phagocytophilum, and most likely also B. miyamotoi (Eisen et al., 2016; Krause et al., 2015; Lane et al., 1994; Merten and Durden, 2000; Teglas and Foley, 2006).
Lyme borreliosis is the most commonly reported vector-borne disease in the United States. In recent years, state and local health departments have reported more than 30,000 cases annually, which is 3 times the number of cases reported each year in the early 1990’s (Mead, 2015). Anaplasmosis and babesiosis case counts have also increased over recent years, though these diseases are not as common as Lyme borreliosis (Adams et al., 2016; CDC, 2016a, 2016b, 2016c). In 2016, state and local health departments reported more than 4000 cases of anaplasmosis and 1910 cases of babesiosis (CDC, 2016b).
There is substantial overlap among the areas that report Lyme borreliosis, anaplasmosis, and babesiosis. Based on cases reported between 2008 and 2015, fourteen states in the Northeast, mid-Atlantic and upper Midwest regions have been classified as high Lyme borreliosis incidence states (Schwartz et al., 2017). In 2016, these 14 states reported more than 96% of Lyme disease, anaplasmosis and babesiosis cases (CDC, 2016b). Although it is not associated with a notifiable condition, Borrelia miyamotoi was recently recognized as another tick-borne human pathogen in the United States. The spirochete appears to have a broad geographic distribution, and clinicians have documented human cases in northeastern and midwestern states (Chowdri et al., 2013; Crowder et al., 2014; Gugliotta et al., 2013; Jobe et al., 2016; Krause et al., 2015, 2013; Molloy et al., 2015; Nelder et al., 2016). Given that the etiologic agents of Lyme disease, Borrelia miyamotoi disease, anaplasmosis, and babesiosis are sympatric in several regions, it is not surprising that researchers have observed evidence of co-infection with two or more of these agents in both ticks and humans (Barbour et al., 2009; Belongia et al., 1999; Fiorito et al., 2017; Johnson et al., 2017; Krause et al., 2002, 2014; Pritt et al., 2016b). Investigators and public health officials need assays that reliably detect a range of pathogens associated with I. scapularis and I. pacificus in field-collected ticks, including ticks infected with multiple pathogens, to estimate the geographic distribution of pathogens and to assess geographic variation in their prevalence.
Hojgaard et al. (2014b) described a testing algorithm employing paired TaqMan real-time polymerase chain reaction (PCR) assays to detect B. burgdorferi s.s., A. phagocytophilum, and Ba. microti in I. scapularis. The algorithm incorporated 2 different targets per pathogen as well as an I. scapularis actin target. We subsequently determined that the actin primer-probe set could be used to verify DNA integrity in both I. scapularis and I. pacificus-derived samples (Graham et al., 2016). The algorithm included 2 Borrelia targets: a non-coding segment of the chromosome, “gB31”, which is present in Borrelia species including B. burgdorferi s.s. and B. miyamotoi, and a segment of the flagellin gene (fliD), which is present in B. burgdorferi sensu lato (s.l.) species including burgdorferi s.s., but not in B. miyamotoi. Without a B. miyamotoi-specific target, however, identification of B. miyamotoi in ticks required additional amplification and sequencing. Moreover, the algorithm could not differentiate ticks co-infected with B. burgdorferi s.s. and B. miyamotoi from ticks infected with B. burgdorferi s.s. alone (both tested positive for the gB31 and fliD targets) (Hojgaard et al., 2014b).
Following the recent discovery of B. mayonii, a new member of the B. burgdorferi s.l. complex that causes Lyme borreliosis (Pritt et al., 2016a, 2016b), we found that the 2 multiplex assays were not adequately specific. That is, we could not differentiate B. burgdorferi s.s. from B. mayonii using the gB31 and fliD targets. Additional testing suggested that the algorithm also detected both the gB31 and fliD targets in some, but not all, other B. burgdorferi s.l. species. There are 9 named species within the B. burgdorferi s.l. complex that are present in the United States: Borrelia americana, Borrelia andersonii, Borrelia bissettiae, B. burgdorferi s.s., Borrelia californiensis, Borrelia carolinensis, Borrelia kurtenbachii, Borrelia lanei sp. nov., and B. mayonii (Margos et al., 2017; Pritt et al., 2016b; Schotthoefer and Frost, 2015). Investigators have detected B. burgdorferi s.s. and B. mayonii as well as B. andersonii and B. kurtenbachii in field-collected I. scapularis (Burgdorfer et al., 1982; Hamer et al., 2012; Johnson et al., 1984; Lin et al., 2001; Margos et al., 2010; Pritt et al., 2016a, 2016b). Ixodes pacificus is naturally associated with B. burgdorferi s.s. as well as B. americana, B. bissettiae, B. californiensis, and B. lanei sp. nov. (Fedorova et al., 2014; Lane et al., 2013; Margos et al., 2017, 2016; Postic et al., 2007, 1998; Rudenko et al., 2009). Borrelia burgdorferi s.s. and B. mayonii have been culture confirmed as human pathogens in the United States (Benach et al., 1983; Pritt et al., 2016a, 2016b; Steere et al., 1983). Golovchenko et al. (2016) recently isolated a B. bissettiae strain from a Florida resident, but they could not report specific clinical manifestations of the infection due to a lack of clinical evidence. Borrelia bissettiae and/or other species may emerge as human pathogens in the United States in the future. Therefore, it is advantageous to detect and differentiate the full range of Borrelia in field-collected ticks.
Despite the shortcomings of the 2 multiplex assays described by Hojgaard et al. (2014b), we found that combining sensitive, multiplex, real-time PCR assays incorporating a tick DNA control and multiple targets for each pathogen was an efficient and reliable approach for high-throughput testing of field-collected ticks (Feldman et al., 2015; Johnson et al., 2017; Morshed et al., 2015). We therefore sought to refine and expand our pathogen testing scheme in accordance with the ever-growing complexity of ixodes-borne Borrelia. Our goals included (1) incorporating targets to detect and differentiate the 3 ixodes-borne Borrelia species known to cause human disease in the United States (B. burgdorferi s.s., B. mayonii, and B. miyamotoi) from each other and from other ixodes-borne borreliae, (2) integrating a target that would allow us to detect other Borrelia species for putative identification by sequencing and/or to bank for future testing if new pathogenic Borrelia species emerge or known species are shown to cause human illness, (3) incorporating at least 2 targets per pathogen, and (4) maintaining the ability to verify DNA integrity. While others have developed molecular testing schemes to detect and differentiate multiple pathogens in Ixodes ticks (e.g., Courtney et al., 2004; Dibernardo et al., 2014; Eshoo et al., 2015; Tokarz et al., 2009, 2017; Ullmann et al., 2005; Wroblewski et al., 2017), we know of no published algorithm to detect and differentiate the etiologic agents of Lyme borreliosis (both B. burgdorferi s.s. and B. mayonii), Borrelia miyamotoi disease, anaplasmosis, and babesiosis, and certainly not one that achieves all 4 of our goals.
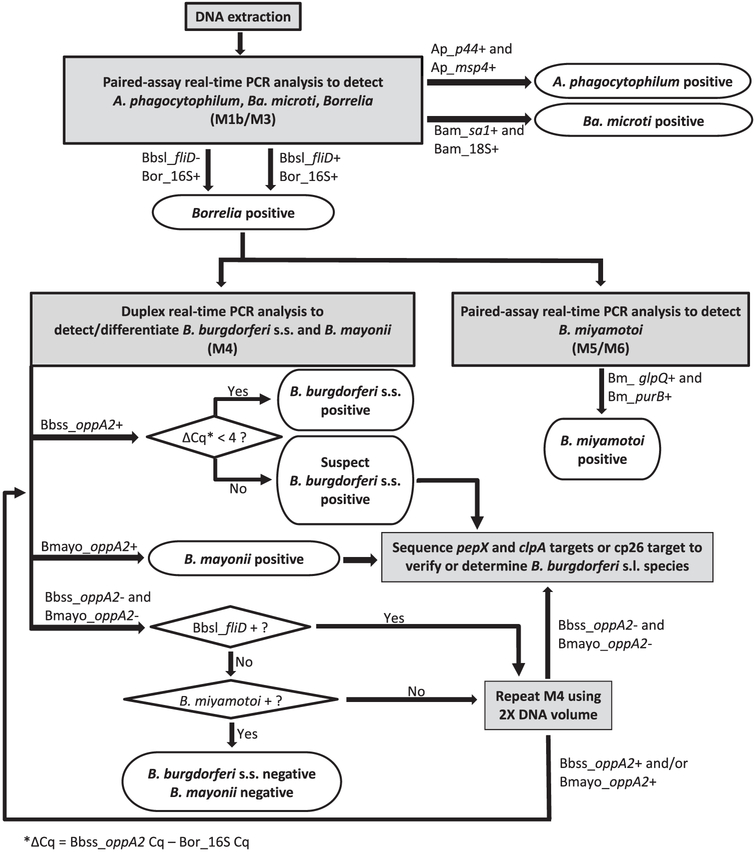
Here we describe an algorithm to detect and differentiate A. phagocytophilum, Ba. microti, B. burgdorferi s.s., B. mayonii, B. miyamotoi, and other Borrelia species in unfed I. scapularis and I. pacificus. The algorithm includes: (1) paired multiplex TaqMan real-time PCR assays for the detection of A. phagocytophilum, Ba. microti, and Borrelia, (2) a duplex TaqMan real-time PCR assay to detect and differentiate B. burgdorferi s.s. and B. mayonii in Borrelia-positive samples, (3) paired TaqMan real-time PCR assays to detect B. miyamotoi in Borrelia-positive samples, and (4) sequencing protocols to putatively identify other Borrelia species, verify B. mayonii positives, and resolve suspect realtime PCR results (Fig. 1). We assessed the algorithm’s sensitivity to each target pathogen, its overall specificity, and its performance on field-collected I. scapularis nymphs from Minnesota.
Fig. 1.
Flow chart for a testing algorithm to detect and differentiate A. phagocytophilum, Ba. microti, B. miyamotoi, B. burgdorferi s.s., B. mayonii, and other B. burdorferi s.l. species in I. scapularis and I. pacificus. Ap, A. phagocytophilum target; Bam. Ba. microti target; Bbsl, B. burgdorferi s.l. target; Bor. pan-Borreha target; Bm, B. miyamotoi target; Bbss, B. burgdorferi s.s. target; Bmayo, B. mayonii target.
2. Materials and methods
2.1. Real-time PCR
Our testing algorithm incorporated 5 real-time PCR assays (Fig. 1). All primer and probe sequences and per-reaction concentrations appear in Table 1. For the first step, in which we sought to detect A. phagocytophilum, Ba. microti, and Borrelia, we modified the paired multiplex TaqMan real-time PCR assays described by Hojgaard et al. (2014b). The modified assays incorporated a pan-Borrelia primer-probe set adapted from Parola et al. (2011) as well as adjustments to reaction volume, some primer concentrations, cycling conditions, and the quantitation cycle (Cq) determination mode. The first assay targeted segments of the genes encoding flagellin (fliD) in B. burgdorferi s.s. and B. mayonii, and the genes encoding A. phagocytophilum P44 outer membrane proteins (p44), and Ba. microti secreted antigen 1 (sa1). For simplicity, we hereafter refer to the fliD target as “B. burgdorferi s.l. fliD” because the assay detected this target in multiple B. burgdorferi s.l. species including both B. burgdorferi s.l. species known to cause human disease in the United States. The first assay also included a tick actin target to verify the integrity of DNA samples derived from I. scapularis and I. pacificus. Hereafter, we refer to this assay as “M1b”, because it incorporates the same targets as the assay previously termed “M1” (Hojgaard et al., 2014b) with modified fliD primer concentrations. The second assay targeted Borrelia 16S rDNA, the genes encoding A. phagocytophilum major surface protein 4 (msp4), and Ba. microti 18S rDNA. We developed this assay to replace “M2” (Hojgaard et al., 2014b). As it incorporated a unique combination of targets, we named it “M3.” For both M1b and M3, each 10-μl reaction included 1X iQ Multiplex Powermix (Bio-Rad Laboratories, Hercules, CA, USA), 200 nM each probe, 300–600 nM each primer (see Table 1), and 4.8 μl template. Real-time cycling conditions comprised an initial 3-min activation step at 95 °C followed by 40 cycles of 95 °C for 10 s and 60 °C for 45 s.
Table 1.
Primers and probes used in a testing algorithm designed to detect and differentiate B. burgdorferi s.s., B. mayonii, B. miyamotoi, A. phagocytophilum and Ba. microti in I. scapularis and I. pacificus.
| Target | Primer/Probe | Sequence (5′-3′) | Amplicon size (nt) | Reference | Concentration/Reaction (nM) |
|---|---|---|---|---|---|
| M1b Assay | |||||
| B. burgdorferi s.l. fliDa | fliD-F | TGG TGA CAG AGT GTA TGA TAA TGG AA | 78 | Zeidner et al., 2001 | 400 |
| fliD-R | ACT CCT CCG GAA GCC ACA A | Zeidner et al., 2001 | 400 | ||
| fliD-probe | FAM-TGC TAA AAT GCT AGG AGA TTG TCT GTC GCC-BHQ1 | Zeidner et al., 2001 | 200 | ||
| A. phagocytophilum p44 | p44-Fb | ATG GAA GGT AGT GTT GGT TAT GGT ATT | 77 | Courtney et al., 2004 | 300 |
| p44-Rb | TTG GTC TTG AAG CGC TCG TA | Courtney et al., 2004 | 300 | ||
| p44-probeb | HEX-TGG TGC CAG GGT TGA GCT TGA GAT TG-BHQ1 | Courtney et al., 2004 | 200 | ||
| Ba. microti sa1 | sa1-F | ACA GAA TGC AGT CGG TGA AG | 115 | Hojgaard et al., 2014b | 300 |
| sa1-R | ATC AAG GAG AGT GGA TAG GTT TG | Hojgaard et al., 2014b | 300 | ||
| sa1-probe | CalRd610-CCA TTG ACG CTG TTG TTG CTC ACA-BHQ2 | Hojgaard et al., 2014b | 200 | ||
| I. scapularis and I. pacificus actin gene | actin-F | GCC CTG GAC TCC GAG CAG | 77 | Hojgaard et al., 2014b | 300 |
| actin-R | CCG TCG GGA AGC TCG TAG G | Hojgaard et al., 2014b | 300 | ||
| actin-probe | Quas705-CCA CCG CCG CCT CCT CTT CTT CC-BHQ3 | Hojgaard et al., 2014b | 200 | ||
| M3 Assay | |||||
| Borrelia | 16S-F | AGC YTT TAA AGC TTC GCT TGT AG | 148 | Kingry et al., 2017a | 600 |
| 16S rDNA | 16S-R | GCC TCC CGT AGG AGT CTG G | Kingry et al., 2017a | 600 | |
| 16S-probe | FAM-CCG GCC TGA GAG GGT GAW CGG-BHQ1 | Kingry et al., 2017a | 200 | ||
| A. phagocytophilum msp4 | msp4-F | TAT ATC CAA CTT CAA CTT CCA CTC | 93 | Hojgaard et al., 2014b | 300 |
| msp4-R | CAT TCA AGT TCG CTA AGA GTT TAC | Hojgaard et al., 2014b | 300 | ||
| msp4-probe | HEX-CTC CGC CAA TAG CAT AGC CAG TTG-BHQ1 | Hojgaard et al., 2014a | 200 | ||
| Ba. microti | 18S-F | CGA CTA CGT CCC TGC CCT TTG | 99 | Hojgaard et al., 2014b | 400 |
| 18S rDNA | 18S-R | ACG AAG GAC GAA TCC ACG TTT C | Hojgaard et al., 2014b | 400 | |
| 18S-probe | Quas705-AC ACC GCC CGT CGC TCC TAC CG-BHQ3 | Hojgaard et al., 2014b | 200 | ||
| M4 Assay | |||||
| B. burgdorferi s.s. oppA2 | Bb-F | AAT TTT TGG TTC CAT ACC C | 162 | This study | 450 |
| B. mayonii oppA2 | Bmayo-F | GCC CGA TTT AAT CAA AGA | 144 | This study | 450 |
| B. burgdorferi s.s. oppA2 and B. mayonii oppA2 | Bb/Bmayo-R | CTG TCA ATA GCA AGA GTT AA | This study | 900 | |
| B. burgdorferi s.s. oppA2 | Bb-probe | HEX-CGT TCA ATA CAC ACA TCA AAC CAC T-BHQ1 | This studyc | 200 | |
| B. mayonii oppA2 | Bmayo-probe | FAM-ACA CGC ACA TTA AAC CGC TTG AT-BHQ1 | This studyc | 200 | |
| M5 Assay | |||||
| B. miyamotoi glpQ | BmglpQ-F | GAC CCA GAA ATT GAC ACA ACC ACA A | 108 | Graham et al., 2016d | 600 |
| BmglpQ-R | TGA TTT AAG TTC AGT TAG TGT GAA GTC AGT | Graham et al., 2016d | 600 | ||
| BmglpQ-probe | CalRd610-CAA TCG AGC TAG AGA AAA CGG AAG ATA TTA CG-BHQ2 | Graham et al., 2016d | 200 | ||
| M6 Assay | |||||
| B. miyamotoi purB | BmpurB-F | TCC TCA ATG ATG AAA GCT TTA | 121 | Graham et al., 2016 | 100 |
| BmpurB-R | GGA TCA ACT GTC TCT TTA ATA AAG | Graham et al., 2016 | 100 | ||
| BmpurB-probe | CalRd610-TCG ACT TGC AAT GAT GCA AAA CCT-BHQ2 | Graham et al., 2016 | 200 | ||
| I. scapularis and I. pacificus actin gene | actin-F, actin-R, and actin-probe as in panel M1b | ||||
| Amplification/Sequencing | |||||
| B. burgdorferi s.l. clpA | clpAF1237 | AAA GAT AGA TTT CTT CCA GAC | 982e | Wang et al., 2014 | 500 |
| clpAR2218 | GAA TTT CAT CTA TTA AAA GCT TTC | Wang et al., 2014 | 500 | ||
| clpAF1255 | GAC AAA GCT TTT GAT ATT TTA G | 850e | Margos et al., 2008 | 500 | |
| clpAR2104 | CAA AAA AAA CAT CAA ATT TTC TAT CTC | Margos et al., 2008 | 500 | ||
| B. burgdorferi s.l. pepX | pepXF449 | TTA TTC CAA ACC TTG CAA TCC | 724 | Margos et al., 2008 | 500 |
| pepXR1172 | GTT CCA ATG TCA ATA GTT TC | Margos et al., 2008 | 500 | ||
| pepXF449 | as above | 668 | Margos et al., 2008 | 500 | |
| pepXR1115 | TGT GCC TGA AGG AAC ATT TG | Margos et al., 2008 | 500 | ||
| B. mayonii | Bm_cp26_OF | CTC ATA TCC CTC TCC TTT GAT | 749 | This study | 900 |
| Bmayo_06250 – | Bm_cp26_OR | TCT GGG CAT ATT TCA GTG AT | This study | 900 | |
| Bmayo_06255 (cp26) | Bm_cp26_IF | TTA CAG ACT AGT GAA CAT A | 337 | This study | 900 |
| Bm_cp26_IR | CAA ATA CAT TAA CCA AGG AGC A | This study | 900 |
nt, nucleotides; fliD, flagellin gene; p44, P44 outer membrane protein gene; sa1, secreted antigen 1 gene; msp4, major surface protein 4 gene; oppA2, oligopeptide permease periplasmic A2 gene; glpQ, glycerophosphodiester phosphodiesterase gene; purB, adenylosuccinate lyase gene; clpA, Clp protease subunit A gene; pepX, Dipeptidyl aminopeptidase gene; Bmayo_06250–Bmayo_06255 (cp26), segment of circular plasmid 26 between Bmayo_06250 and Bmayo_06255; CalRd610, CAL Fluor Red 610, FAM, 6-Carboxyfluorescein; HEX, Hexachloro-Fluorescein Phosphoramidite; Quas705, Quasar 705; BHQ1, BHQ2, BHQ3: Black Hole Quenchers 1–3.
The B. burgdorferi s.l. fliD primer/probe set detects multiple B. burgdorferi s.l. species including B. burgdorferi s.s. and B. mayonii.
A. phagocytophilum p44 primers and probe appear elsewhere as “msp2-F, msp2-R, and msp2-P” (Courtney et al., 2004; Hojgaard et al., 2014b). The msp2 gene in Anaplasma marginale is in the same protein superfamily as the p44 genes in A. phagocytophilum. There is, however, a distinct msp2 gene in the A. phagocytophilum genome (Dunning Hotopp et al., 2006; Lin et al., 2004). The A. phagocytophilum primer/probe set in panel M1b detects a conserved A. phagocytophilum p44 gene sequence. We have changed the primer and probe names accordingly.
M4 panel probes were adapted from the TaqMan real-time PCR assay described in Pritt et al. (2016b) Supplemental Materials.
B. miyamotoi glpQ primers and probe adapted from Bacon et al. (2005).
Amplicon size for the clpA target varies slightly among B. burgdorferi s.l. species and strains. We’ve listed the amplicon sizes for B. burgdorferi B31.
To detect and differentiate B. burgdorferi s.s. and B. mayonii in Borrelia-positive samples (Fig. 1), we designed a duplex TaqMan realtime PCR assay targeting the oligopeptide permease periplasmic A2 gene (oppA2). This duplex assay, hereafter “M4,” employed 2 species-specific forward primers (Bb-F and Bmayo-F), a single reverse primer designed to anneal to a conserved segment of the oppA2 sequence (Bb/Bmayo-R), and species-specific probes (Bb-probe, Bmayo-probe). In addition to the 3 primers and 2 probes (Table 1), each 25-μl duplex reaction included 1X iQ Multiplex Powermix (Bio-Rad), and 5–10 μl template. Real-time cycling conditions comprised an initial 3-min activation step at 95 °C followed by 40 cycles of 95 °C for 15 s and 58 °C for 1 min.
To detect B. miyamotoi in Borrelia-positive samples, we incorporated a pair of real-time TaqMan PCR assays into the algorithm, hereafter “M5” and “M6”. These assays are described in detail elsewhere (Graham et al., 2016). Briefly, M5 targeted the gene encoding B. miyamotoi glycerophosphodiester phosphodiesterase (glpQ). M6 targeted the adenylosuccinate lyase (purB) gene and the tick actin target included in M1b (Table 1). Each 10-μl reaction included 1X iQ Multiplex Powermix (Bio-Rad), 200 nM probe, 600 nM each primer, and 4.8 μl template. Real-time cycling conditions were identical to those for assays M1b and M3.
We used CFX Manager 3.1 software (Bio-Rad) with the Cq determination mode set to regression to analyze results for all real-time PCR assays.
2.2. Sequencing
We integrated a set of 3 amplification and sequencing protocols into the algorithm to (1) putatively identify Borrelia-positive samples that test negative for B. burgdorferi s.s., B. mayonii, and B. miyamotoi, (2) putatively identify B. burgdorferi s.l. fliD-positive samples that test negative for B. burgdorferi s.s. and B. mayonii, (3) verify B. mayonii-positive samples, and (4) confirm the presence of B. burgdorferi s.s. in samples with suspect real-time PCR results (Fig. 1). All 3 protocols employed (semi-)nested PCR to facilitate specific amplification of potentially scarce Borrelia DNA from tick material. Primer sequences and per-reaction concentrations appear in Table 1.
To identify borreliae in ticks infected with a single B. burgdorferi s.l. species, we amplified and sequenced segments of the housekeeping genes encoding Clp protease subunit A (clpA) or Dipeptidyl aminopeptidase (pepX) as described at pubMLST (http://pubmlst.org/borrelia/ ; Margos et al., 2015) and elsewhere (Margos et al., 2008; Wang et al., 2014) with minor modifications. Briefly, each 25-μl outer reaction included 1X HotStar Taq Master Mix (Qiagen, Valencia, CA, USA), primers clpAF1237 and clpAR2218 or pepXF449 and pepXR1172, 0.5 μl 25mM MgCl2 to bring the final concentration to 2mM MgCl2, and 5–10 μl template. Touchdown cycling conditions for outer reactions were as described at PubMLST, but with annealing temperatures of 60 °C-52 °C for the first set of pepX amplification cycles and 52 °C for the second set of pepX amplification cycles as suggested by Wang et al. (2014). Each 50-μl inner reaction included 1X HotStar Taq Master Mix (Qiagen), inner primers clpAF1255 and clpAR2104 or pepX449 and pepXR1115, 1 μl 25mM MgCl2 to bring the final concentration to 2mM MgCl2, and 5–10 μl of the outer reaction product. Cycling conditions were as described at PubMLST, but with a 15 min initial activation at 95 °C and an annealing temperature of 52 °C for pepX.
Because the clpA and pepX primers generate amplicons from all B. burgdorferi s.l. species, they do not allow for verification of B. mayonii in samples that are co-infected with another B. burgdorferi s.l. species, e.g., B. burgdorferi s.s. We therefore developed an amplification and sequencing protocol targeting a B. mayonii-specific segment of circular plasmid 26 (cp26) between Bmayo_06250 and Bmayo_06255 (Kingry et al., 2016). Each 25-μl outer reaction and each 50-μl inner reaction included 1X HotStar Taq Master Mix (Qiagen), 900 nM each primer (outer reaction: Bm_cp26_OF, Bm_cp26_OR; inner reaction: Bm_cp26_IF, Bm_cp26_IR), 0.5 μl 25mM MgCl2 to bring the final MgCl2 concentration to 2mM per reaction, and 5–10 μl DNA (outer reaction) or 5–10 μl outer reaction product (inner reaction). Cycling conditions included a 15 min initial activation at 95 °C followed by 30 (outer reaction) or 40 (inner reaction) cycles of 94 °C for 30 s, 56 °C (outer reaction) or 55 °C (inner reaction) for 30 s and 72 °C for 1 min, and a final 10 min extension at 72 °C.
Before sequencing, we visualized inner products on a 1% agarose gel to verify the presence of an approximately 850-nucleotide (nt) (clpA), 668-nt (pepX) or 337-nt (cp26) amplicon. The remaining product was purified using the QIAquick PCR Purification Kit (Qiagen). We sequenced each product using the inner amplification primers and BigDye Terminator v3.1 Ready Reaction Mix, removed unincorporated dyes with the BigDye Xterminator Kit (ThermoFisher Scientific Inc., Waltman, MA, USA), and analyzed the samples on an ABI 3130XL genetic analyzer. Using Lasergene 12 software (DNASTAR, Madison, WI, USA), we aligned forward and reverse sequences to generate a consensus sequence with at least 2-fold coverage of every nt. We manually trimmed poor or ambiguous sequence and any primer sequence from either end of the consensus sequence. We then used the Basic Local Alignment Search Tool (BLAST) to identify similar sequences in GenBank. We also queried the PubMSLT database using the clpA and pepX consensus sequences to identify similar alleles.
2.3. Analytical sensitivity and specificity
2.3.1. Pathogen DNA
All Borrelia DNA used in this study came from reference collections maintained in the Bacterial Diseases Branch of the Division of Vector-Borne Diseases, Centers for Disease Control and Prevention (DVBD, CDC, Fort Collins, CO, USA), and had been extracted from cultured isolates (Supplemental Table 1). DNA from A. phagocytophilum USG3, cultured in HL-60 human promyeloblast cells, was provided by the Rickettsial Zoonoses Branch, DVBD, CDC (Atlanta, GA, USA). Babesia microti DNA, extracted from a human blood sample (reference number 1953), was provided by the Division of Parasitic Diseases and Malaria, CDC, in accordance with the CDC Human Subjects Research Protocol, “Use of residual diagnostic specimens from humans for laboratory methods research” (Atlanta, GA, USA).
2.3.2. Tick DNA
To extract DNA from individual I. scapularis nymphs, we homogenized individual ticks with ≈545 mg 2.0mm yttria-stabilized zirconium oxide beads (Glen Mills, Clifton, NJ, USA) in 470 μl lysis mix comprised of buffer ATL, 20 μl proteinase K, and 0.5% DX anti-foaming reagent (Qiagen). We disrupted the sample for a total of 2 min using a Mini-Beadbeater-96 (BioSpec Products, Bartlesville, OK, USA) as described in Graham et al. (2016), then incubated the homogenate for 10–12 min at 56 °C. After centrifuging the sample for 30 s at 1000 x g, we processed 200 μl using the QIAcube HT automated nucleic acid isolation system and the cador Pathogen 96 QIAcube HT Kit (Qiagen). Following lysis with buffer VXL and the addition of buffer ACB, the instrument loaded 650 μl sample into the capture plate and applied a 3 min vacuum at 35 kPa. The column was washed with 600 μl buffer AW1 and subjected to a 2 min vacuum at 35 kPa. Subsequent wash and dry steps followed the cador Pathogen 96 QIAcube HT Kit V3 program (Qiagen). At the final step, DNA was eluted by adding 100 μl buffer AVE to the column, incubating for 2 min, and applying a 55 kPa vacuum for 6 min.
To prepare tick DNA for use in spiking reactions, we homogenized pooled, colony-reared I. scapularis nymphs (DVBD, Fort Collins) in 180 μl buffer ATL with 0.5% DX anti-foaming reagent (Qiagen) for 2 min in a Mini-Beadbeater-96 (BioSpec) with ≈545 mg 2.0mm yttria-stabilized zirconium oxide beads (GlenMills). We then added 20 μl proteinase K and incubated the homogenate overnight at 56 °C before extracting DNA using the DNeasy Blood and Tissue Kit (Qiagen).
2.3.3. Recombinant plasmids
To assess the sensitivity of the paired assays (M1b and M3) that provide the foundation for our testing algorithm (Fig. 1), we constructed 6 recombinant plasmids. We used the M1b and M3 primers (Table 1) to amplify the fliD and 16S targets from B. burgdorferi B31, the p44 and msp4 targets from A. phagocytophilum USG3, and the sa1 and 18S targets from Ba. microti. We purified each product from a 2% agarose gel using Freeze ‘N Squeeze DNA extraction columns (Bio-Rad). Each purified product was then cloned it into a pCR4-TOPO plasmid vector and propagated in TOP10 chemically competent E. coli using the TOPO TA Cloning Kit for Sequencing (ThermoFisher). We verified that each recombinant plasmid contained the correct insert by preparing forward and reverse sequence reactions using BigDye Terminator v3.1 Ready Reaction Mix and pCR4-TOPO plasmid vector T3 and T7 primers (ThermoFisher). We removed unincorporated dyes and analyzed each sequence as in Section 2.2. We subsequently linearized plasmids containing each complete target sequence using restriction enzyme NotI and purified the linearized plasmid DNA using the QIAquick PCR Purification Kit (Qiagen). Samples with an absorption ratio (A260/A280) < 1.79 were re-purified by ethanol precipitation.
2.3.4. Limit of detection for each real-time PCR target
We determined the double-stranded DNA concentration of each plasmid or genomic DNA template using a Qubit 2.0 fluorometer and the Qubit dsDNA HS Assay Kit (ThermoFisher) immediately before preparing serial dilutions for limit of detection (LOD) experiments. We included 5 replicates at each concentration, and each reaction also contained 6 ng I. scapularis DNA, which is the average amount of double-stranded DNA in 5 μl extract prepared from a single nymph as described in Section 2.3.2. For all LOD experiments, primer-probe sets were run in multiplex. Unless otherwise noted, we conducted LOD testing for each target on 2 different days, using fresh DNA dilutions each day, for a total of 10 replicates per concentration per target, and we defined the LOD for each target as the lowest concentration for which all 10 replicates yielded positive PCR results (target Cq value < 40).
To determine the copy-number LOD for each M1b and M3 target, we prepared serial dilutions of the linearized recombinant plasmids containing each target to achieve the equivalent of 3, 6, or 10 target copies per reaction. To ascertain whether the Borrelia targets in M1b and M3 showed similar sensitivity to B. burgdorferi s.s., B. mayonii, and B. miyamotoi, we also determined the LOD for both targets using genomic DNA from these 3 species. Specifically, we determined the B. burgdorferi s.l. fliD LOD using B. burgdorferi B31 and B. mayonii MN14-1539 genomic DNA, and the Borrelia 16S LOD using genomic DNA from these 2 strains as well as B. miyamotoi CT13-2396. We prepared serial dilutions to achieve the equivalent of 3, 6, 10, 20, 30, or 40 genomes per reaction. Each reaction also included 6 ng I. scapularis DNA. We based our genome copy-number calculations on estimated genome sizes for B. burgdorferi B31 (1.52 Mbp; NCBI Genome Database), B. mayonii MN14-1539 (1.30 Mbp; Kingry et al., 2016), and B. miyamotoi CT13-2396 (1.28 Mbp; Kingry et al., 2017b). Because our A. phagocytophilum and Ba. microti genomic DNA stocks contained host DNA, we determined genomic DNA LODs empirically for A. phagocytophilum and Ba. microti by preparing 2-fold serial dilutions to achieve the equivalent of 25y–400 fg/reaction and identifying the lowest concentration at which both multiplex panels (M1b and M3) detected 10 replicates. We defined the M1b/M3 paired-assay LOD for each pathogen as the LOD associated with the less sensitive target.
Using the same method we used to determine genomic LODs for the Borrelia targets in M1b and M3, we used genomic DNA from B. burgdorferi s.s. strains B31 and MN88-0003 and B. mayonii strain MN14-1539 to determine the LODs for the B. burgdorferi s.s. and B. mayonii oppA2 targets in M4. We previously determined the LOD for each B. miyamotoi target in assays M5 and M6 using similar methods (Graham et al., 2016).
2.3.5. Impact of increased tick DNA or co-infection on assay sensitivity
To assess the impact of increased tick DNA concentrations (> 6 ng/reaction) on M1b/M3 sensitivity, we prepared reactions containing genomic DNA from B. burgdorferi s.s., A. phagocytophilum, or Ba. microti at the M1b/M3 paired-assay LOD and I. scapularis DNA at each of 4 concentrations between 12 and 96 ng/reaction (Table 4). To assess the impact of co-infecting pathogens on the M1b/M3 LOD for B. burgdorferi s.s., A. phagocytophilum and Ba. microti, we prepared reactions containing 6 ng I. scapularis DNA, genomic DNA from a single pathogen at the paired-assay LOD, and genomic DNA from a second pathogen at 10, 102 and 103 times its LOD.
Table 4.
Number of replicates in which assays M1b and M3 detected each target in genomic DNA at the paired-assay limit of detection (LOD; 10 fg B. burgdorferi s.s., 200 fg A. phagocytophilum, 200 fg Ba. microti) in the presence of increasing I. scapularis tick DNA.
|
I. scapularis DNA (ng/reaction) |
M1b |
M3 |
||||
|---|---|---|---|---|---|---|
| Bbsl_fliD | Ap_p44 | Bam_sa1 | Bor_16S | Ap_msp4 | Bam_18S | |
| 12 | 10/10 | 10/10 | 9/10 | 10/10 | 10/10 | 9/10 |
| 24 | 5/10 | 10/10 | 7/10 | 10/10 | 10/10 | 10/10 |
| 48 | 4/10 | 10/10 | 9/10 | 10/10 | 10/10 | 10/10 |
| 96 | 2/10 | 9/10 | 7/10 | 10/10 | 10/10 | 10/10 |
Bbsl, Borrelia burgdorferi s.l. target; Bor, pan-Borrelia target; Ap, A. phagocytophilum target; Bam, Ba. microti target.
To assess the sensitivity of assay M4 to B. burgdorferi s.s. and B. mayonii in co-infected samples, we prepared reactions containing I. scapularis DNA, genomic B. mayonii DNA at concentrations equivalent to 10, 102, 103, or 104 genomes per reaction, and B. burgdorferi s.s. DNA at concentrations equivalent to 10, 102, 103, or 104 genomes per reaction. We included 5 replicates at each B. mayonii concentration in the presence of B. burgdorferi s.s. at each concentration (Table 5).
Table 5.
Number of replicates in which assay M4 detected the B. burgdorferi s.s. oppA2 and B. mayonii oppA2 targets in reactions spiked with genomic DNA from both pathogens at concentrations equivalent to 10–104 genomes per reaction.
| B. mayonii genomes | 10 B. burgdorferi s.s. genomes |
102
B. burgdorferi s.s. genomes |
103
B. burgdorferi s.s. genomes |
104
B. burgdorferi s.s. genomes |
||||
|---|---|---|---|---|---|---|---|---|
| Bbss_oppA2 | Bmayo_oppA2 | Bbss_oppA2 | Bmayo_oppA2 | Bbss_oppA2 | Bmayo_oppA2 | Bbss_oppA2 | Bmayo_oppA2 | |
| 10 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 3/5 | 5/5 | 0/5 |
| 102 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 |
| 103 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 |
| 104 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 |
2.3.6. Algorithm specificity
We assessed the algorithm’s ability to differentiate Borrelia species using a panel of 20 Borrelia strains comprising B. burgdorferi s.s., B. mayonii, B. miyamotoi, and 6 other Borrelia species (Supplemental Table 1). Using the M1b, M3, M4, M5, and M6 assays, we screened I. scapularis DNA-spiked stocks of each strain at concentrations of approximately 10 pg and 50 fg Borrelia DNA per reaction. We subsequently amplified and sequenced the clpA and pepX targets from all Borrelia species that tested negative for B. burgdorferi s.s., B. mayonii, and B. miyamotoi to determine if we could reliably identify other B. burgdorferi s.l. species.
2.4. Pathogen detection in field-collected ticks
To further assess assay performance, we tested DNA extracted as described in Section 2.3.2 from 192 individual I. scapularis nymphs collected by drag sampling on public lands in Clearwater County (n=115) and Hubbard County (n=77) in Minnesota in June 2015. Each set of extractions included at least 1 tick-free extraction control for every 18 field-collected samples. In addition to extraction controls, each M1b/M3 real-time PCR run included at least 1 no-template control and a positive control comprised of pooled recombinant plasmid DNA containing each pathogen target at a concentration equivalent to approximately 20 copies per reaction. To assess the integrity of each field-collected tick extract, we analyzed the distribution of the I. scapularis actin Cq values generated by all 192 samples following M1b/M3 testing. Specifically, we used JMP 11 statistical software (v. 11.1.1 SAS Institute, Inc., 2013) to construct a boxplot, and we identified outliers with an I. scapularis Cq value > the upper whisker value (3rd quartile +1.5[interquartile range]). We re-tested these outliers, as well as any samples that yielded an I. scapularis actin curve with an end relative fluorescent unit (RFU) value < 400. If the I. scapularis actin results still indicated insufficient or poor quality DNA upon repeat, we prepared a second extract from 200 μl of the remaining tick triturate. If the I. scapularis actin results for the second extract did not indicate adequate DNA quantity and quality, we excluded the tick from infection prevalence analyses. We also repeated and/or prepared a second extract from samples yielding inconsistent M1b/M3 results (e.g., A. phagocytophilum p44-positive/msp4-negative).
Each M4 real-time run included at least 1 negative control for every 18 Borrelia-positive samples and a positive control containing genomic B. burgdorferi s.s. and B. mayonii DNA at concentrations equivalent to approximately 20 genome copies each per reaction. Each M5/M6 realtime run included at least 1 negative control for every 18 Borrelia-positive test samples and a positive control containing the appropriate recombinant plasmid at a concentration equivalent to approximately 10 target copies per reaction.
We used PooledInfRate, Version 4.0 (Biggerstaff, 2009) to calculate 95% confidence intervals for infection rates with each pathogen.
3. Results and discussion
Here we describe the development and evaluation of an algorithm for high throughput testing of field-collected, host-seeking I. scapularis and I. pacificus for 5 tick-borne human pathogens.
3.1. Algorithm sensitivity
3.1.1. Algorithm limits of detection for Anaplasma phagocytophilum and Babesia microti
The LOD associated with each pathogen target in the algorithm appears in Table 2. Because the M1b and M3 assays each included 1 of 2 targets for A. phagocytophilum and Ba. microti, and because a sample must test positive for both targets to be considered positive for the pathogen (Fig. 1), we identified the algorithm LOD for each of these pathogens as the LOD associated with the target that was less sensitive to genomic pathogen DNA. The algorithm LOD for A. phagocytophilum was 200 fg of our genomic A. phagocytophilum stock, which our results suggest was approximately equivalent to 3 copies of the msp4 target (Table 2). The A. phagocytophilum genome contains a single copy of the msp4 gene (Dunning Hotopp et al., 2006). Assuming a single genome per bacterium, we would thus expect the algorithm to reliably detect as few as 3 A. phagocytophilum organisms per reaction. Notably, the A. phagocytophilum genome contains as many as 80 full-length or reserve/silent p44 genes, although the specific p44 gene family complement varies between strains (Dunning Hotopp et al., 2006). This explains why the p44 target was much more sensitive to genomic A. phagocytophilum DNA than the msp4 target even though the p44 target copy LOD was higher.
Table 2.
The limit of detection (LOD) for each pathogen target in the algorithm, defined as the lowest target copy or genome copy number consistently detected by real-time PCR. We determined each target copy number LOD using linearized recombinant DNA from a plasmid containing the target sequence. We determined each pathogen LOD using genomic DNA. Borrelia DNA was extracted from spirochete cultures and genome concentrations were estimated as described in Section 2.3.4. Because our A. phagocytophilum and Ba. microti genomic DNA stocks contained host DNA, we could not extrapolate a genome equivalent of the genomic DNA LOD.
| Target (assay) | Target Copy LOD |
Target copies per genome |
Pathogen LOD (fg DNA or Borrelia spp. genomes) |
|
|---|---|---|---|---|
| species | LOD | |||
| A. phagocytophilum p44 (M1b) | 20 copies | multiple | A. phagocytophilum | 25 fg |
| A. phagocytophilum msp4 (M3) | 3 copies | 1 | A. phagocytophilum | 200 fg* |
| Ba. microti sa1 (M1b) | 3 copies | 1 | Ba. microti | 200 fg* |
| Ba. microti 18S (M3) | 6 copies | 2 | Ba. microti | 100 fg |
| B. burgdorferi s.l. fliD (M1b) | 6 copies | 1 | B. burgdorferi s.s. | 6 genomes |
| B. mayonii | 30 genomes | |||
| Borrelia 16S (M3) | 3 copies | 1 | B. burgdorferi s.s. | 3 genomes |
| B. mayonii | 6 genomes | |||
| B. miyamotoi | 3 genomes | |||
| B. burgdorferi s.s. oppA2 (M4) | NT | B. burgdorferi s.s | 3–6 genomes* ,a | |
| B. mayonii oppA2 (M4) | NT | B. mayonii | 10 genomes* ,b | |
| B. miyamotoi glpQ (M5) | 5 copiesc | 1 | B. miyamotoi | 5 genomesc‘* |
| B. miyamotoi purB (M6) | 5 copiesc | 1 | B. miyamotoi | 5 genomesc |
NT, not tested.
Indicates algorithm LOD for each pathogen.
The M4 assay consistently detected B. burgdorferi s.s. strain B31 down to the equivalent of 3 genomes, and B. burgdorferi s.s. strain MN88-0003 down to the equivalent of 6 genomes.
Borrelia-positive samples that initially test negative for B. mayonii oppA2 can be retested using 2X the standard DNA volume to bring the algorithm LOD for B. mayonii down to ≤ 5 genomes per 1X sample volume.
We found that M3 was slightly more sensitive to genomic Ba. microti DNA than M1b (Table 2). This is likely because there are 2 copies of the 18S target in the Ba. microti genome (Cornillot et al., 2012), and there is a single copy of the sa1 target in the Ba. microti genome, as indicated by BLAST analysis of the sa1 target site (JX112361.1:714-828) against the GenBank database, which contains the complete Ba. microti strain RI genome (Cornillot et al., 2012). The algorithm LOD for Ba. microti was 200 fg of our Ba. microti genomic stock, which our results suggest was equivalent to approximately 3 copies of the sa1 target, or 3 Ba. microti genomes (Table 2). Therefore, assuming haploid form, we expect the algorithm to reliably detect 3 Ba. microti organisms per reaction. Souza et al. (2016) reported that, used in singleplex, the Ba. microti sa1 and 18S primer-probe sets consistently detected the equivalent of 14 and 12 parasites per reaction, although they observed detection down to the equivalent of 2.4 and 0.5 parasites, respectively, in DNA extracted from parasitic hamster blood. Babesia microti is haploid in mammalian hosts (Goethert and Telford, 2014), so these results suggest that in a diagnostic context, a singleplex assay targeting sa1 (1 copy/genome) would reliably detect as few as 14 genomes, and a singleplex assay targeting 18S (2 copies/genome) would reliably detect as few as 24 genomes. It is possible that these singleplex real-time PCR assays are in fact less sensitive to Ba. microti in DNA extracted from blood than M1b and M3 are to Ba. microti DNA in a tick extract. We acknowledge, however, that the true LOD for each target in the algorithm, in terms of actual organisms per reaction, is likely slightly higher than reported here, as the DNA extraction process is unlikely to yield 100% of the pathogen DNA present in a sample, regardless of the matrix.
3.1.2. M1b/M3 limit of detection for B. burgdorferi s.s., B. mayonii, and B. miyamotoi
The pan-Borrelia 16S target LOD was between 3 and 6 genomes in representative strains of 3 human-disease causing Borrelia (Table 2). Assuming a single genome per spirochete, we would therefore expect M3 to reliably detect 6 or fewer B. burgdorferi s.s., B. mayonii or B. miyamotoi spirochetes per reaction. Our results for the fliD target, however, indicate that M1b has an LOD of 6 B. burgdorferi s.s. spirochetes and 30 B. mayonii spirochetes (Table 2). This may be due to flagellin gene sequence variations between these species. The fliD target sequence is well-conserved among B. burgdorferi s.s. strains; BLAST analysis of the 78-nt sequence from strain B31 (CP017201.1:149547-149704), the strain from which the fliD primer and probe sequences were derived, indicated 98.7% identity (1 nt difference) with B. burgdorferi s.s. strain JD1, and 100% identity with all other B. burgdorferi s.s. strains for which homologous sequence was available in the GenBank database (strains PAbe, PAli, CA382, N40, and ZS7). The fliD target sequence in B. mayonii (CP015780.1: 150092-150169) differed from the target sequence in B. burgdorferi B31 by 4 nt. Notably, however, mismatches between the fliD primer sequences and the primer-annealing sequences in B. mayonii fell at least 10 nt from the 3′ end of each primer. Because M1b might fail to detect B. mayonii at very low concentrations, we determined that all Borrelia 16S-positive samples – whether they are positive or negative for fliD – should undergo additional testing for B. mayonii, B. burgdorferi s.s., and B. miyamotoi (Fig. 1). Given this caveat, we concluded that the M1b/M3 assays had a combined LOD of ≤ 6 spirochetes for all 3 target Borrelia species. We treated 10 fg B. burgdorferi B31 DNA (equivalent to approximately 6 genomes) as the M1b/M3 Borrelia species LOD for all subsequent testing.
While the Borrelia 16S results determine the need for further testing with assays M4, M5 and M6, we kept the B. burgdorferi s.l. fliD target in M1b so that the first phase of testing would include two targets for B. burgdorferi s.s., the primary agent of the most commonly-reported vector-borne disease in the United States. In most cases, fliD also serves as a second target for B. mayonii. In addition, the B. burgdorferi s.l. fliD results are used to determine if additional testing is warranted for Borrelia 16S-positives samples that test negative for both B. burgdorferi s.s. and B. mayonii (Fig. 1).
3.1.3. Impact of co-infection or increased tick DNA on the limit of detection for each M1b/M3 pathogen target in multiplex
All M1b and M3 targets reliably detected genomic DNA over at least 5 logs (LOD–104 x LOD). Preferential amplification of one target over another can impact pathogen detection using multiplex PCR, particularly when one target is highly abundant and a second target is at relatively low abundance (Bustin et al., 2009; Elnifro et al., 2000). To assess the impact of co-infecting pathogens on M1b and M3 sensitivity, we evaluated detection of genomic DNA from each pathogen at its M1b/M3 LOD in the presence of genomic DNA from a second pathogen at 10–103 times its LOD. The results are summarized in Table 3. Borrelia burgdorferi s.l. fliD and Ba. microti sa1 were the only 2 targets for which we observed decreased sensitivity in the presence of other pathogen DNA.
Table 3.
Number of replicates in which assays M1b and M3 detected each target in genomic DNA at the paired-assay limit of detection (LOD; 10 fg B. burgdorferi s.s., 200 fg A. phagocytophilum, 200 fg Ba. microti) in the presence of DNA from a second pathogen at 10–103 x its LOD.
| Second pathogen |
B. burgdorferi s.s. (10 fg) |
A. phagocytophilum (200 fg) |
Ba. microti (200 fg) |
|||
|---|---|---|---|---|---|---|
| fliD (M1b) | 16S (M3) | p44 (M1b) | msp4 (M3) | sa1 (M1b) | 18S (M3) | |
| 2 pg A. phagocytophilum | 9/10 | 10/10 | n/a | n/a | 9/10 | 10/10 |
| 20 pg A. phagocytophilum | 7/10 | 10/10 | n/a | n/a | 7/10 | 10/10 |
| 200 pg A. phagocytophilum | 8/10 | 10/10 | n/a | n/a | 9/10 | 10/10 |
| 2 pg Ba. microti | 9/10 | 10/10 | 10/10 | 10/10 | n/a | n/a |
| 20 pg Ba. microti | 7/10 | 10/10 | 10/10 | 10/10 | n/a | n/a |
| 200 pg Ba. microti | 8/10 | 10/10 | 10/10 | 10/10 | n/a | n/a |
| 100 fg B. burgdorferi s.s. | n/a | n/a | 10/10 | 10/10 | 9/10 | 10/10 |
| 1 pg B. burgdorferi s.s. | n/a | n/a | 10/10 | 9/10 | 8/10 | 10/10 |
| 10 pg B. burgdorferi s.s. | n/a | n/a | 10/10 | 10/10 | 9/10 | 10/10 |
Since we determined the analytical sensitivity of each assay using reactions spiked with the amount of tick DNA we would expect to have in 5 μl extract prepared from a single nymph using our standard DNA extraction protocol (6 ng), we also assessed the impact of increasing the amount of tick DNA on sensitivity. Such a scenario could occur in extracts from adult ticks, extracts prepared using an alternative extraction method that produces a more concentrated eluate, or extracts from pooled ticks. The addition of 12 to 96 ng genomic tick DNA simulated the use of DNA extracted as in Section 2.3.2 from pools of 2 to 16 nymphs. The results are summarized in Table 4. Increasing the amount of tick DNA did not appear to impact detection using any of the M3 panel targets. Two of the M1b targets, however, Ba. microti sa1, and – more dramatically – B. burgdorferi s.l. fliD, lost some sensitivity as the tick DNA concentration increased above 12 ng/reaction. The sa1 and fliD targets were likely less sensitive because the M1b assay, unlike the M3 assay, included a tick actin target; amplification of the actin target from increasingly abundant tick DNA may have impacted detection of the sa1 and fliD targets in scarce pathogen DNA. It may also be that both co-infection and additional tick DNA impacted detection of B. burgdorferi s.l. fliD and Ba. microti sa1 in multiplex because we were testing for detection at the combined assay LOD, which was equivalent to the target LOD for both fliD and sa1, whereas targets for the same pathogens in M3, (Borrelia 16S and Ba. microti 18S respectively), were slightly more sensitive (Table 2).
We previously observed similar amplification of the tick actin target from I. scapularis and I. pacificus DNA (Graham et al., 2016). Given that I. scapularis appeared to impact assay sensitivity to some pathogen targets multiplexed with the tick actin target in M1b, likely because preferential amplification of the tick target hindered amplification of scarce pathogen DNA, we would expect I. pacificus DNA to similarly impact M1b sensitivity to B. burgdorferi s.l. and Ba. microti. We would not expect I. pacificus DNA to have any more impact than I. scapularis DNA on the sensitivity of assays that do not include a tick target. Therefore, though we conducted all testing using I. scapularis DNA, we would expect the algorithm to yield similar results and to be subject to similar limitations for I. pacificus-derived DNA.
3.1.4. Re-testing samples that yield ambiguous M1b/M3 results using B. burgdorferi s.l fliD and Ba. microti sa1 in singleplex
One advantage of using paired real-time assays for pathogen detection is that if host material and/or co-infecting pathogen DNA in-hibits detection of a scarce pathogen target in one panel, we may still pick it up with the other panel. Based on the results of our LOD experiments, the algorithm specifies that a sample that tests negative for B. burgdorferi s.l. fliD and positive for Borrelia 16S, or negative for Ba. microti sa1 and positive for 18S, should be re-tested using M3 along with B. burgdorferi s.l. fliD or Ba. microti sa1 in singleplex.
Run in singleplex, we detected B. burgdorferi s.l. fliD in 10 fg B. burgdorferi s.s. DNA in at least 9 of 10 replicates in the presence of up to 96 ng I. scapularis DNA, up to 200 pg A. phagocytophilum DNA, and up to 200 pg Ba. microti DNA. While it is true that a Borrelia 16S-positive sample should undergo additional testing for B. burgdorferi s.s., B. mayonii and B. miyamotoi whether it tests positive for B. burgdorferi s.l. fliD or not (Fig. 1), it is helpful to determine if a sample is B. burgdorferi s.l. fliD positive or negative with as much confidence as possible. As noted in Section 3.1.2, these results are used to determine if additional testing is needed for Borrelia-positives samples that test negative for both B. burgdorferi s.s. and B. mayonii (Fig. 1).
Run in singleplex, we consistently detected Ba. microti sa1 in 200 fg Ba. microti DNA in at least 9 of 10 replicates in the presence of up to 48 ng I. scapularis DNA, and in 8 of 10 replicates in the presence of 96 ng I. scapularis DNA. Experiments did not indicate that sa1 run in singleplex detected 200 fg Ba. microti in the presence of co-infecting pathogens more consistently than sa1 run in M1b multiplex. We therefore acknowledge some loss of algorithm sensitivity to Ba. microti in co-infected samples.
3.1.5. Algorithm limit of detection for Borrelia miyamotoi
The algorithm specifies that samples that test positive for Borrelia 16S using the paired M1b and M3 assays should subsequently undergo testing for B. burgdorferi s.s., B. mayonii, and B. miyamotoi (Fig. 1). We incorporated paired TaqMan real-time PCR assays M5 and M6 to test for B. miyamotoi in all Borrelia-positive samples. Previous characterization of this assay revealed that it reliably detected ≤ 5 genomes, or 5 spirochetes, per reaction, did not detect B. burgdorferi s.s., B. turicatae or B. lonestari, and was unlikely to detect other relapsing fever Borrelia (Graham et al., 2016). Since we determined that Borrelia 16S had an LOD of 3 B. miyamotoi genomes (Table 2) in this study, we conclude that the algorithm provides specific detection of B. miyamotoi down to at least 5 spirochetes per reaction.
3.1.6. Algorithm limit of detection for Borrelia burgdorferi s.s. and B. mayonii in ticks infected with one or both pathogens
A tick that tests positive for Borrelia 16S might contain DNA from B. burgdorferi s.s., B. mayonii, or both; co-infected ticks accounted for more than 30% of the B. mayonii-positive ticks identified by Pritt et al. (2016b). Therefore, we developed an assay, M4, that could detect and distinguish B. burgdorferi s.s. and B. mayonii in ticks carrying one or both species. Using genomic DNA, we determined that this duplex assay had a B. burgdorferi s.s. LOD of ≤ 6 genomes (Table 2). M4 was slightly less sensitive to B. mayonii, reliably detecting DNA down to the equivalent of 10 genomes. Thus, a sample yielding between 6 and 10 B. mayonii genomes per reaction, the equivalent of between 6 and 10 spirochetes per reaction, would be expected to test positive for Borrelia 16S, and it might test negative for B. burgdorferi s.l. fliD and B. mayonii oppA2 (Table 2). If it also tested negative for both B. burgdorferi s.s. and B. miyamotoi, 2X sample volume would be re-tested using M4 (Fig. 1), which should reliably detect B. mayonii at concentrations of ≤ 5 genomes, or 5 spirochetes, per 1X sample volume. Given this caveat, and given that the Borrelia 16S LOD was 3 genomes for B. burgdorferi s.s. and 6 genomes for B. mayonii (Table 2), we conclude that the algorithm has an overall LOD of ≤ 6 spirochetes for both B. burgdorferi s.s. and B. mayonii.
In reactions spiked with tick DNA and both B. burgdorferi s.s. and B. mayonii, M4 reliably detected B. burgdorferi s.s. at concentrations as low as 10 genomes per reactions in the presence of up to 104 B. mayonii genomes. It also reliably detected the equivalent of 10 B. mayonii genomes in the presence of up to 100 B. burgdorferi s.s. genomes (Table 5). Although M4 failed to detect 10 genomes B. mayonii in the presence of 103–104 genomes B. burgdorferi s.s. DNA, the assay consistently detected the B. mayonii oppA2 target when B. mayonii was present at concentrations ≥ 100 genomes per reaction (Table 5). Nonetheless, we acknowledge that the algorithm may fail to detect B. mayonii at very low concentrations in the presence of relatively abundant B. burgdorferi s.s. DNA.
3.2. Algorithm specificity
3.2.1. Real-time PCR specificity for Borrelia species
To assess the algorithm’s specificity for B. burgdorferi s.s., B. mayonii, and B. miyamotoi, specifically its ability to differentiate these 3 species from each other and from other tick-borne borreliae, we tested high and low concentrations (10 pg and 50 fg per reaction) of the 20 Borrelia strains in Supplemental Table 1. Table 6 shows the real-time PCR results for each strain at both concentrations. The algorithm detected and correctly identified the 5 B. burgdorferi s.s., 2 B. mayonii and 4 B. miyamotoi strains we tested. As expected, the M3 assay detected the Borrelia 16S target in all Borrelia with similar Cq values for all 104 fg replicates (range: 24.15–25.18) and all 50 fg replicates (range: 31.16–32.49), suggesting similar sensitivity across species and strains. As expected, all B. miyamotoi strains were negative for B. burgdorferi s.l. fliD. M1b detected the fliD target in all B. burgdorferi s.s. and B. mayonii strains, with Cq values suggesting similar sensitivity across all B. burgdorferi s.s. strains, and slightly less sensitivity to B. mayonii, consistent with the findings from our LOD experiments (Table 2). The M1b assay showed a wide range of sensitivity, however, to all other B. burgdorferi s.l. strains. M1b detection of the fliD target in B. bissettiae, B. californiensis, and B. kurtenbachii was comparable to detection of the fliD target in the B. burgdorferi s.s. strains. The assay detected the fliD target only in the high concentration replicates of B. americana and 1 B. andersonii strain, and the fliD Cq values were well above the Borrelia fliD Cq values associated with B. burgdorferi s.s. and B. mayonii strains at the same concentration. Three strains tested negative for fliD at both the high and low concentrations. These findings confirmed the need for additional testing to differentiate Borrelia-positive samples.
Table 6.
Real-time PCR results (Cq values) for each Borrelia target included in our testing algorithm for B. burgdorferi s.l. and B. miyamotoi strains run at high (104 fg/reaction) and low (50 fg/ reaction) concentrations. Cq values<40 were considered positive. A target that failed to amplify within 40 cycles was considered negative (−). The difference between the Borrelia 16S (Bor_16S) and B. burgdorferi s.s. oppA2 (Bbss_oppA2) Cq values was calculated for each Bbss_oppA2-positive replicate to allow for differentiation between true (ΔCq < 4) and false (ΔCq > 4) B. burgdorferi s.s. positives. .
| Borrelia species | Strain | fg DNA per reaction |
Bbsl_fliD Cq (M1b) |
Bor_16S Cq (M3) |
Bmiya_purB Cq (M5) |
Bmiya_qlpQ Cq (M6) |
Bmayo_oppA2 Cq (M4) |
Bbss_oppA2 Cq (M4) |
ΔCq (Bbss_oppA2–Bor_16S) |
|---|---|---|---|---|---|---|---|---|---|
| B. burgdorferi s.s. | B31T | 104 | 25.14 | 24.56 | – | – | – | 25.88 | 1.32 |
| 50 | 32.29 | 31.49 | – | – | – | 33.27 | 1.78 | ||
| B. burgdorferi s.s. | IRS | 104 | 25.15 | 24.41 | – | – | – | 26.01 | 1.60 |
| 50 | 32.61 | 31.61 | – | – | – | 33.94 | 2.33 | ||
| B. burgdorferi s.s. | CA6 | 104 | 24.87 | 24.54 | – | – | – | 25.38 | 0.84 |
| 50 | 32.72 | 31.19 | – | – | – | 32.51 | 1.32 | ||
| B. burgdorferi s.s. | MN88-0003 | 104 | 24.86 | 24.89 | – | – | – | 26.16 | 1.27 |
| 50 | 31.94 | 31.87 | – | – | – | 33.38 | 1.51 | ||
| B. burgdorferi s.s. | SI1 | 104 | 25.01 | 24.77 | – | – | – | 26.23 | 1.46 |
| 50 | 31.92 | 31.19 | – | – | – | 34.68 | 3.49 | ||
| B. mayonii | MN14—1420T | 104 | 26.39 | 25.08 | – | – | 26.21 | – | n/a |
| 50 | 32.80 | 31.16 | – | – | 33.55 | – | n/a | ||
| B. mayonii | MN14-1539 | 104 | 26.64 | 25.18 | – | – | 26.20 | – | n/a |
| 50 | 32.91 | 32.00 | – | – | 34.84 | – | n/a | ||
| B. miyamotoi | HT31T | 104 | – | 24.59 | 22.88 | 25.69 | – | – | n/a |
| 50 | – | 31.62 | 30.70 | 33.20 | – | – | n/a | ||
| B. miyamotoi | HT24 | 104 | – | 24.99 | 23.59 | 26.27 | – | – | n/a |
| 50 | – | 32.04 | 30.85 | 34.55 | – | – | n/a | ||
| B. miyamotoi | CT13-2396 | 104 | – | 24.94 | 23.49 | 25.76 | – | – | n/a |
| 50 | – | 31.55 | 30.61 | 34.50 | – | – | n/a | ||
| B. miyamotoi | RI13-2395 | 104 | – | 25.00 | 23.60 | 26.22 | – | – | n/a |
| 50 | – | 31.82 | 31.08 | 34.85 | – | – | n/a | ||
| B. americana | SCW-41; | 104 | 30.87 | 24.92 | – | – | – | 31.11 | 6.19 |
| subgroup AT | 50 | 32.49 | n/a | ||||||
| B. andersonii | 21038 | 104 | – | 25.05 | – | – | – | – | n/a |
| 50 | – | 31.89 | – | – | – | – | n/a | ||
| B. andersonii | SI-10 | 104 | 32.76 | 24.15 | – | – | – | – | n/a |
| 50 | – | 31.16 | – | – | – | – | n/a | ||
| B. bissettiae | DN127T | 104 | 24.79 | 24.65 | – | – | – | – | n/a |
| 50 | 32.20 | 31.40 | – | – | – | – | n/a | ||
| B. bissettiae | CA389 | 104 | 24.81 | 24.53 | – | – | – | – | n/a |
| 50 | 31.86 | 31.66 | – | – | – | – | n/a | ||
| B. californiensis | CA20 | 104 | 25.53 | 24.92 | – | – | – | – | n/a |
| 50 | 32.16 | 31.83 | – | – | – | – | n/a | ||
| B. carolinensis | SCW-22 | 104 | – | 25.10 | – | – | – | – | n/a |
| 50 | – | 31.93 | – | – | – | – | n/a | ||
| B. kurtenbachii | 25015T | 104 | 25.38 | 24.79 | – | – | – | 38.23 | 13.44 |
| 50 | 32.71 | 32.00 | – | – | – | – | n/a | ||
| B. kurtenbachii | IL96-255 | 104 | 25.10 | 24.72 | – | – | – | 35.64 | 10.92 |
| 50 | 31.87 | 31.42 | – | – | – | – | n/a |
Bbsl, Borrelia burgdorferi s.l. target; Bbss, Borrelia burgdorferi s.s. target; Bor, pan-Borrelia target; Bmiya, B. miyamotoi target; Bmayo, B. mayonii target.
denotes Type Strain.
Given that the B. miyamotoi purB and glpQ genes are not present in B. burgdorferi s.l. spirochetes (Pettersson et al., 2007; Schwan et al., 2003), it was not surprising that M5 and M6 clearly differentiated the 4 B. miyamotoi strains from all B. burgdorferi s.l. strains (Table 6). M4 correctly identified all B. burgdorferi s.s. and B. mayonii strains. We also observed poor detection of the B. burgdorferi s.s. oppA2 target, however, in B. americana and B. kurtenbachii at high concentrations. The false B. burgdorferi s.s. oppA2 positive Cq values were more than 6 cycles higher than the Borrelia 16S Cq values associated with the same strains at the same concentration, while the difference between B. burgdorferi s.s. oppA2 and Borrelia 16S Cq values for the 5 B. burgdorferi s.s. strains was consistently less than 4 at both high and low concentrations (Table 6). The algorithm therefore specifies that a sample testing positive for B. burgdorferi s.s. using the M4 assay but with a B. burgdorferi s.s. oppA2 Cq value more than 4 cycles above the sample’s Borrelia 16S Cq value should be considered suspect, and the Borrelia species identification should be verified by sequencing (Fig. 1). While we recognize that this complicates the algorithm, we note that false B. burgdorferi s.s. oppA2 positives are likely to be very rare. Assuming spirochete loads per nymph are similar across B. burgdorferi s.l. species, we would expect the spirochete load in the vast majority of naturally-infected nymphs to be less than 300,000 spirochetes (Barbour et al., 2009). We use approximately 2% of a nymph per reaction, so a single reaction would very rarely contain 6000 B. burgdorferi s.l. genomes, which is roughly equivalent to 10 pg B. burgdorferi s.l. DNA, the high concentration in our specificity experiment.
3.2.2. Using clpA and pepX sequences to resolve Borrelia burgdorferi s.l species
In keeping with the algorithm, we amplified and sequenced the clpA and pepX targets from 9 B. burgdorferi s.l. strains: the 6 that tested negative for B. miyamotoi, B. mayonii and B. burgdorferi s.s., and the 3 that would be considered suspect based on their discordant B. burgdorferi s.s. oppA2 and Borrelia 16S Cq values. We have deposited clpA and pepX sequences for strains B. americana SCW-41, B. andersonii 21038 and SI-10, B. californiensis CA20, and B. carolinensis SCW-22 in GenBank (accession numbers MF582579–MF582588). Target sequences for all other strains were already available. We were able to correctly identify all 9 strains from our test panel based on the clpA and pepX sequences. Our results indicated that clpA and pepX sequence analysis, using both the GenBank and MLST databases, should allow us to assign a sample to a particular species with reasonable confidence when both sequences show > 97% similarity to strains from a single species, and when one or both sequences show ≤ 97% identity with all other species. Analysis of the clpA and pepX sequences was also sufficient to verify B. mayonii positives, as indicated by the results from our field-collected tick testing (Section 3.3).
Overall, the algorithm detected and correctly identified all of the human disease causing Borrelia strains in our test panel, and it did not mistake any of the B. burdorferi s.l. species not known to be associated with human disease in the United States for B. burgdorferi s.s. or B. mayonii.
3.3. Pathogen detection in field-collected ticks
To assess the algorithm’s performance on field-collected samples, we tested 192 host-seeking I. scapularis nymphs collected in Clearwater and Hubbard Counties in Minnesota in June 2015. Based on the I. scapularis actin Cq values associated with repeated testing of 1 nymph from Hubbard County, we determined that we could not reliably detect pathogen DNA in this specimen. This nymph was therefore excluded from all further analyses. Table 7 shows the infection prevalence with our 5 target pathogens: B. burgdorferi s.s., B. mayonii, B. miyamotoi, A. phagocytophilum, and Ba. microti.
Table 7.
Prevalence of each of the five pathogens targeted by our testing algorithm in I. scapularis nymphs collected in June 2015 on public lands in two north-central Minnesota counties, and prevalence of ticks testing positive for two or more of these pathogens.
| Collection Site | Total no. nymphs |
No. positive for each pathogena (infection rate; 95% CI (%)) |
No. positive for multiple pathogensa (infection rate; 95% CI (%)) |
||||
|---|---|---|---|---|---|---|---|
|
Borrelia burgdorferi s.s. |
Borrelia mayonii |
Borrelia miyamotoi |
Anaplasma phagocytophilum |
Babesia microti | |||
| Clearwater | 115 | 36 (31.3; 23.4–40.2) | 1 (0.9; 0.0–4.1) | 0 (0.0; 0.0–3.2) | 14 (12.2; 7.1–19.1) | 13 (11.3; 6.5–18.1) | 15 (13.0; 7.8–20.1) |
| Hubbard | 76 | 31 (40.8; 30.2–52.0) | 3 (3.9; 1.0–10.3) | 2 (2.6; 0.5–8.3) | 6 (7.9; 3.3–15.6) | 3 (3.9; 1.0–10.3) | 9 (11.8; 6.0–20.6) |
| Total | 191 | 67 (35.1; 28.6–42.0) | 4 (2.1; 0.7–4.9) | 2 (1.0; 0.2–3.4) | 20 (10.5; 6.7–15.4) | 16 (8.4; 5.0–13.0) | 24 (12.6; 8.4–17.8) |
No., number; CI, confidence interval.
If we detected two or more target pathogens in a tick, that tick is included in the “No. positive” for each pathogen for which it was positive. It is also included in the count of ticks positive for multiple pathogens.
Hubbard and Clearwater are adjacent counties in north-central Minnesota. We chose to use I. scapularis nymphs from these counties to assess algorithm performance on field-collected samples because Clearwater is one of the few counties in which B. mayonii-infected I. scapularis nymphs had been previously documented (Pritt et al., 2016b), and because we also expected to detect B. burgdorferi s.s., A. phagocytophilum and Ba. microti in nymphs from this region. In the period between 2008 and 2011, both counties reported human cases of Lyme borreliosis and human anaplasmosis (> 50 cases/100,000 residents), and between 2004 and 2011, both counties also reported a small number of babesiosis cases (Robinson et al., 2015). Though these data reflect the county of residence associated with each case and not necessarily the county of exposure, Robinson et al. (2015) found that residents of these and other rural counties were more likely to have been exposed in their county of residence than residents of more urban counties.
Our B. mayonii results (Table 7) are consistent with the only previously published data for B. mayonii infection in I. scapularis nymphs from Minnesota. Pritt et al. (2016b) detected B. mayonii in 1 of 59 nymphs (1.7%; 95% CI: 0.1–7.9%) collected in Clearwater County between April and July 2015, the period within which our ticks were also collected. The algorithm also yielded B. burgdorferi s.s., B. miyamotoi, and A. phagocytophilum infection prevalence rates (Table 7) within the ranges reported for the prevalence of each pathogen in I. scapularis nymphs from the upper Midwest. Barbour et al. (2009) detected B. burgdorferi s.s. in 6.4–41.0% and B. miyamotoi in 0–8.2% of I. scapularis nymphs collected between mid-May and late August in the years 2004 through 2007 from each of 21 different sites in Minnesota, Michigan and Wisconsin. Murphy et al. (2017) detected A. phagocytophilum in 0.0–17.7% of questing I. scapularis nymphs collected by drag sampling at each of 24 sites is Wisconsin in 2015. There is limited data available on Ba. microti prevalence in questing I. scapularis nymphs collected in the Midwest. Stromdahl et al. (2014) detected Ba. microti in 3.3% of 215 I. scapularis nymphs submitted between 2002 and 2012 from soldiers at a Minnesota military installation. The algorithm yielded a higher Ba. microti infection prevalence in the nymphs we tested (8.4%), but the lower end of the 95% confidence interval (5.0%) falls within the 95% confidence interval associated with the Stromdahl et al. (2014) data (1.5–6.3%).
The algorithm detected co-infections in 24 ticks. Nine (38%) tested positive for B. burgdorferi s.s. and A. phagocytophilum, and 7 (29%) tested positive for B. burgdorferi s.s. and Ba. microti. Four ticks were infected with B. burgdorferi s.s., A. phagocytophilum and Ba. microti, and 1 tick was infected with B. burgdorferi s.s., Ba. microti, and B. mayonii. We also detected co-infections with B. mayonii and B. miyamotoi, B. burgdorferi s.s. and B. miyamotoi, and B. burgdorferi s.s. and B. mayonii (1 each).
Notably, though the algorithm includes a number of steps to clarify ambiguous real-time PCR results (Fig. 1), we determined the pathogen infection status for 185 of the 192 field-collected ticks (96%) based on a single round of real-time PCR testing using assays M1b and M3 followed by a single round of testing using assays M4, M5 and M6 to identify Borrelia-positive samples to species. All samples that tested positive for the Borrelia 16S target also tested positive for the B. burgdorferi s.l. fliD target, including the 2 B. miyamotoi-positive ticks, because 1 was coinfected with B. burgdorferi s.s. and the other with B. mayonii. All samples that tested positive for Ba. microti 18S also tested positive for Ba. microti sa1, so we did not need to conduct any follow-up testing using B. burgdorferi s.l. fliD or Ba. microti sa1 in singleplex. Two samples initially yielded inconsistent results for the 2 A. phagocytophilum targets. One tested negative for both targets upon repeat and was therefore negative for A. phagocytophilum by the algorithm. The second sample yielded inconsistent A. phagocytophilum results again upon repeat, so we prepared a second DNA extract from the leftover triturate. We detected both A. phagocytophilum targets in the second extract and classified the sample as A. phagocytophilum positive.
The B. burgdorferi s.s. oppA2 Cq values were within 4 of the Borrelia 16S Cq values for all samples that tested positive for B. burgdorferi s.s. We verified the presence of B. mayonii DNA in 2 samples by sequencing the clpA and pepX targets. The other 2 B. mayonii-positive samples were co-infected with B. burgdorferi s.s. We verified the presence of B. mayonii in these samples by amplifying and sequencing the cp26 target. BLAST analysis confirmed that all sequences were identical to the homologous regions in B. mayonii MN14-1420 and MN14-1539.
Two Borrelia-positive samples that tested positive for both the B. burgdorferi s.l. fliD target and the Borrelia 16S target subsequently tested negative for B. miyamotoi, B. burgdorferi s.s., and B. mayonii. We repeated M4 testing using 10 μl of each sample, which revealed that 1 of the 2 samples was positive for B. burgdorferi s.s. We note that though M4 initially failed to detect B. burgdorferi s.s. in this sample, the algorithm prompted additional testing that ultimately allowed us to identify this human disease causing Borrelia. The second Borrelia-positive sample continued to test negative for B. burgdorferi s.s. and B. mayonii. BLAST analysis of the clpA sequence from this sample revealed 99% identity with B. kurtenbachii strains, 99% identity with a single B. bissettiae strain, and ≤ 96% identity with all other strains and species. The most similar allele in the pubMLST database was clpA 127, with which it shared 576/579 nts (99% identity). The pepX sequence was identical to the homologous region in B. kurtenbachii strain IL96-255, 99% similar to other B. kurtenbachii strains in GenBank, and ≤ 96% similar to available sequences for all other species. It contained pepX allele 107. The clpA 127 and pepX 107 alleles were associated exclusively with B. kurtenbachii isolates in pubMLST. We therefore concluded that this nymph was not infected with B. burgdorferi s.s., B. mayonii, or B. miyamotoi, and that it was most likely positive for B. kurtenbachii.
4. Conclusion
We conclude that the algorithm described here can reliably detect and distinguish 5 known human pathogens associated with I. scapularis. A 2-panel multiplex Taqman assay (M1b/M3) provides the foundation for the algorithm and detects A. phagocytophilum, Ba. microti, B. burgdorferi s.s., B. miyamotoi and B. mayonii at concentrations as low as 3–6 genomes per reaction. Subsequent testing with assays M4, M5, and M6 allows detection and differentiation of human disease causing Borrelia in Borrelia-positive samples, while amplification and sequencing protocols enable further verification and identification of other Borrelia species (Fig. 1). The algorithm integrates multiple targets for each pathogen with a tick actin target that can be used to verify the integrity of I. scapularis and I. pacificus DNA extracts. The inclusion of a pan-Borrelia target means that DNA samples testing positive for Borrelia but negative for B. miyamotoi, B. burgdorferi s.s. and B. mayonii can be putatively identified by sequencing and/or banked to allow for future screening for Borrelia species that have not yet been identified as human disease agents. We validated the algorithm using nymphal tick DNA because most Lyme borreliosis, anaplasmosis, and babesiosis cases occur during peak nymphal host-seeking activity, indicating that the host-seeking nymphal stage poses a particular risk to human health (Eisen et al., 2017). Based on our assessment of algorithm sensitivity in the presence of increased tick DNA, however, we believe that this testing scheme would also be useful for testing individual or pooled larvae or adult ticks. This scheme could also be used to test blood-fed I. scapularis and I. pacificus, although we do not know how the presence of blood in field-collected specimens might impact assay sensitivity. The algorithm will be useful for testing I. scapularis and I. pacificus collected throughout the United States in support of surveillance and research programs that assess the risk of human exposure to the agents of Lyme borreliosis, Borrelia miyamotoi disease, anaplasmosis, and babesiosis.
Supplementary Material
Acknowledgements
We thank Drs. William Nicholson and Sandor Karpathy of the Rickettsial Zoonoses Branch of the DVBD, CDC, for providing A. phagocytophilum DNA, and Maniphet Xayavong of the Parasitic Diseases Branch of the Division of Parasitic Diseases and Malaria, CDC for providing Ba. microti DNA. The B. californiensis DNA used in this study came from the Lane Tick and Tick/Vertebrate DNA Extract Collection, generously donated to the Bacterial Disease Branch of the DVBD, CDC by Dr. Robert S. Lane. The following individuals kindly provided and/or prepared DNA from all other Borrelia strains used in this study: Dr. Jeannine Petersen, Dr. Martin Schriefer, Dr. Luke Kingry, Dr. Robert Gilmore, Dr. Claudia Molins, Adam Replogle, and Mark Pilgard, all of the Bacterial Diseases Branch, DVBD, CDC. Drs. Kingry and Petersen also provided helpful comments on the manuscript. We thank Nicole Breuner for providing all of the colony-reared ticks used in this study, Dr. Tammi Johnson for providing field-collected ticks for assay evaluation, and Janae Stovall and Karen Boroughs for technical assistance with sequencing.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ttbdis.2017.12.005.
References
- Adams DA, Thomas KR, Jajosky RA, Foster L, Sharp P, Onweh DH, Schley AW, Anderson WJ, 2016. Summary of notifiable infectious diseases and conditions – United States, 2014. MMWR Morb. Mortal. Wkly. Rep 63, 1–152. [DOI] [PubMed] [Google Scholar]
- Bacon RM, Pilgard MA, Johnson BJ, Piesman J, Biggerstaff BJ, Quintana M, 2005. Rapid detection methods and prevalence estimation for Borrelia lonestari glpQ in Amblyomma americanum (Acari: Ixodidae) pools of unequal size. Vector-Borne Zoon. Dis 5, 146–156. [DOI] [PubMed] [Google Scholar]
- Bakken JS, Dumler S, 2008. Human granulocytic anaplasmosis. Inf. Dis. Clin. N. Am 22, 433–448. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Bunikis J, Travinsky B, Hoen AG, Diuk-Wasser MA, Fish D, Tsao JI, 2009. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am. J. Trop. Med. Hyg 81, 1120–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia EA, Reed KD, Mitchell PD, Chyou PH, Mueller-Rizner N, Finkel MF, Schriefer ME, 1999. Clinical and epidemiological features of early Lyme disease and human granulocytic ehrlichiosis in Wisconsin. Clin. Infect. Dis 29, 1472–1477. [DOI] [PubMed] [Google Scholar]
- Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Habicht GS, Bast TF, Cameron DJ, Ziegler JL, Barbour AG, Burgdorfer W, Edelman R, Kaslow RA, 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med 308, 740–742. [DOI] [PubMed] [Google Scholar]
- Biggerstaff BJ, 2009. PooledInfRate, Version 4.0: a Microsoft® Office Excel© Add-in to Compute Prevalence Estimates from Pooled Samples. Centers for Disease Control and Prevention, Fort Collins, CO, U.S.A. [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP, 1982. Lyme disease-a tick-borne spirochetosis? Science 216, 1317–1319. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- CDC, 2016a. Annual cases of anaplasmosis in the United States, https://www.cdc.gov/anaplasmosis/stats/index.html. (Accessed 28 June 2017).
- CDC, 2016b. National notifiable infectious diseases and conditions: United States. Annual data for 2016. Tables 1, 2c, 2e, and 2i. https://wonder.cdc.gov/nndss/nndss_annual_tables_menu.asp. (Accessed 11 July 2017).
- CDC, 2016c. Number of reported cases of babesiosis, by year, 2014. https://www.cdc.gov/parasites/babesiosis/data-statistics/graphs/graphs.html. (Accessed 28 June 2017).
- Chowdri HR, Gugliotta JL, Berardi VP, Goethert HK, Molloy PJ, Sterling SL, Telford SR, 2013. Borrelia miyamotoi infection presenting as human granulocytic anaplasmosis: a case report. Ann. Int. Med 159, 21–27. [DOI] [PubMed] [Google Scholar]
- Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, Vacherie B, Augagneur Y, Bres V, Duclos A, Randazzo S, Carcy B, Debierre-Grockiego F, Delbecq S, Moubri-Menage K, Shams-Eldin H, Usmani-Brown S, Bringaud F, Wincker P, Vivares CP, Schwarz RT, Schetters TP, Krause PJ, Gorenflot A, Berry V, Barbe V, Ben Mamoun C, 2012. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 40, 9102–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney JW, Kostelnik LM, Zeidner NS, Massung RF, 2004. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. J. Clin. Microbiol 42, 3164–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder CD, Carolan HE, Rounds MA, Honig V, Mothes B, Haag H, Nolte O, Luft BJ, Grubhoffer L, Ecker DJ, Schutzer SE, Eshoo MW, 2014. Prevalence of Borrelia miyamotoi in Ixodes ticks in Europe and the United States. Emerg. Infect. Dis 20, 1678–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibernardo A, Cote T, Ogden NH, Lindsay LR, 2014. The prevalence of Borrelia miyamotoi infection, and co -infections with other Borrelia spp. in Ixodes scapularis ticks collected in Canada. Parasites Vectors 7 10.1186/1756-3305-7-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan MC, Hojgaard A, Hoxmeier JC, Replogle AJ, Respicio-Kingry LB, Sexton C, Williams MA, Pritt BS, Schriefer ME, Eisen L, 2016. Vector competence of the blacklegged tick, Ixodes scapularis, for the recently recognized Lyme borreliosis spirochete Candidatus Borrelia mayonii. Ticks Tick Borne Dis. 7, 665–669. [DOI] [PubMed] [Google Scholar]
- Dolan MC, Piesman J, Mbow ML, Maupin GO, Peter O, Brossard M, Golde WT, 1998. Vector competence of Ixodes scapularis and Ixodes ricinus (Acari: Ixodidae) for three genospecies of Borrelia burgdorferi. J. Med. Entomol 35, 465–470. [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H, 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2 (2), e21 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, 2010. Update on Powassan virus: emergence of a North American tick-borne flavivirus. Annu. Rev. Entomol 55, 95–110. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Eisen L, Beard CB, 2016. County-scale distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the continental United States. J. Med. Entomol 53, 349–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Kugeler KJ, Eisen L, Beard CB, Paddock CD, 2017. Tick-borne zoonoses in the United States: persistent and emerging threats to human health. ILAR J. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE, 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev 13, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshoo MW, Carolan HE, Massire C, Chou DM, Crowder CD, Rounds MA, Phillipson CA, Schutzer SE, Ecker DJ, 2015. Survey of Ixodes pacificus ticks in California reveals a diversity of microorganisms and a novel and widespread ana- plasmataceae species. PLoS One 10 (9), e0135828 10.1371/journal.pone.0135828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N, Kleinjan JE, James D, Hui LT, Peeters H, Lane RS, 2014. Remarkable diversity of tick or mammalian-associated borreliae in the metropolitan San Francisco Bay Area, California. Ticks Tick Borne Dis. 5, 951–961. [DOI] [PubMed] [Google Scholar]
- Feldman KA, Connally NP, Hojgaard A, Jones EH, White JL, Hinckley AF, 2015. Abundance and infection rates of Ixodes scapularis nymphs collected from residential properties in Lyme disease-endemic areas of Connecticut, Maryland, and New York. J. Vector Ecol 40, 198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito TM, Reece R, Flanigan TP, Silverblatt FJ, 2017. Borrelia miyamotoi polymerase chain reaction positivity on a tick-borne disease panel in an endemic region of Rhode Island: a case series. Infect. Dis. Clin. Pract 25, 250–254. [Google Scholar]
- Goethert HK, Telford SR 3rd, 2014. Not out of Nantucket: Babesia microti in southern New England comprises at least two major populations. Parasite Vectors 7, 546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovchenko M, Vancova M, Clark K, Oliver JH Jr., Grubhoffer L, Rudenko N, 2016. A divergent spirochete strain isolated from a resident of the southeastern United States was identified by multilocus sequence typing as Borrelia bissettii. Parasites Vectors 9 10.1186/s13071-016-1353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CB, Pilgard MA, Maes SE, Hojgaard A, Eisen RJ, 2016. Paired real-time PCR assays for detection of Borrelia miyamotoi in North American Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae). Ticks Tick Borne Dis. 7, 1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugliotta JL, Goethert HK, Berardi VP, Telford SR 3rd, 2013. Meningoencephalitis from Borrelia miyamotoi in an immunocompromised patient. N. Engl. J. Med 368, 240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer SA, Hickling GJ, Keith R, Sidge JL, Walker ED, Tsao JI, 2012. Associations of passerine birds, rabbits, and ticks with Borrelia miyamotoi and Borrelia andersonii in Michigan, U.S.A. Parasites Vectors 5 10.1186/1756-3305-5-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojgaard A, Lukacik G, Piesman J, 2014a. Corrigendum to detection of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti, with two different multiplex PCR assays. Ticks Tick Borne Dis. 5, 349–351. [DOI] [PubMed] [Google Scholar]
- Hojgaard A, Lukacik G, Piesman J, 2014b. Detection of Borrelia burgdorferi, Anaplasma phagocytophilum and Babesia microti: with two different multiplex PCR assays. Ticks Tick Borne Dis. 5, 349–351. [DOI] [PubMed] [Google Scholar]
- Jobe DA, Lovrich SD, Oldenburg DG, Kowalski TJ, Callister SM, 2016. Borrelia miyamotoi infection in patients from upper Midwestern United States, 2014–2015. Emerg. Infect. Dis. 22, 1471–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DK, Schiffman EK, Davis JP, Neitzel DF, Sloan LM, Nicholson WL, Fritsche TR, Steward CR, Ray JA, Miller TK, Feist MA, Uphoff TS, Franson JJ, Livermore AL, Deedon AK, Theel ES, Pritt BS, 2015. Human infection with Ehrlichia muris-like pathogen, United States, 2007–2013. Emerg. Infect. Dis 21, 1794–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Schmid GP, Hyde FW, Steigerwalt AG, Brenner DJ, 1984. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int. J. Syst. Bacteriol 34, 496–497. [Google Scholar]
- Johnson TL, Graham CB, Boegler KA, Cherry CC, Maes SE, Pilgard MA, Hojgaard A, Buttke DE, Eisen RJ, 2017. Prevalence and diversity of tick-borne pathogens in nymphal Ixodes scapularis (Acari: Ixodidae) in eastern national parks. J. Med. Entomol 54, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpathy SE, Allerdice ME, Sheth M, Dasch GA, Levin ML, 2016. Co-feeding transmission of the Ehrlichia muris-like agent to mice (Mus musculus). Vector-Borne Zoon. Dis 16, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry LC, Batra D, Replogle A, Rowe LA, Pritt BS, Petersen JM, 2016. Whole genome sequence and comparative genomics of the novel Lyme borreliosis causing pathogen, Borrelia mayonii. PLoS One 11 (12), e0168994 10.1371/journal.pone.0168994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry LC, Anacker M, Pritt B, Bjork J, Respicio-Kingry L, Liu G, Sheldon S, Boxrud D, Strain A, Oatman S, Berry J, Sloan L, Mead P, Neitzel D, Kugeler KJ, Petersen JM, 2017a. Surveillance for and discovery of Borrelia species in U.S. patients suspected of tickborne illness. Clin. Infect. Dis (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingry LC, Replogle A, Batra D, Rowe LA, Sexton C, Dolan M, Connally N, Petersen JM, Schriefer ME, 2017b. Toward a complete North American Borrelia miyamotoi genome. Genome Announc. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PJ, Fish D, Narasimhan S, Barbour AG, 2015. Borrelia miyamotoi infection in nature and in humans. Clin. Microbiol. Infec 21, 631–639 e01557-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PJ, McKay K, Thompson CA, Sikand VK, Lentz R, Lepore T, Closter L, Christianson D, Telford SR, Persing D, Radolf JD, Spielman A, Deer-Associated Infection Study Group, 2002. Disease-specific diagnosis of coinfecting tickborne zoonoses: babesiosis, human granulocytic ehrlichiosis, and Lyme disease. Clin. Infect. Dis 34, 1184–91. [DOI] [PubMed] [Google Scholar]
- Krause PJ, Narasimhan S, Wormser GP, Barbour AG, Platonov AE, Brancato J, Lepore T, Dardick K, Mamula M, Rollend L, Steeves TK, Diuk-Wasser M, Usmani-Brown S, Williamson P, Sarksyan DS, Fikrig E, Fish D, Tick Borne Diseases, G., 2014. Borrelia miyamotoi sensu lato seroreactivity and seroprevalence in the northeastern United States. Emerg. Infect. Dis 20, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PJ, Narasimhan S, Wormser GP, Rollend L, Fikrig E, Lepore T, Barbour A, Fish D, 2013. Human Borrelia miyamotoi infection in the United States. N. Engl. J. Med 368, 291–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane RS, Brown RN, Piesman J, Peavey CA, 1994. Vector competence of Ixodes pacificus and Dermacentor occidentalis (Acari: Ixodidae) for various isolates of Lyme disease spirochetes. J. Med. Entomol 31, 417–424. [DOI] [PubMed] [Google Scholar]
- Lane RS, Fedorova N, Kleinjan JE, Maxwell M, 2013. Eco-epidemiological factors contributing to the low risk of human exposure to ixodid tick-borne borreliae in southern California, USA. Ticks Tick Borne Dis. 4, 377–385. [DOI] [PubMed] [Google Scholar]
- Lin Q, Rikihisa Y, Felek S, Wang X, Massung RF, Woldehiwet Z, 2004. Anaplasma phagocytophilum has a functional msp2 gene that is distinct from p44. Infect. Immun 72, 3883–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Oliver JH Jr., Gao L, Kollars TM Jr., Clark KL, 2001. Genetic heterogeneity of Borrelia burgdorferi sensu lato in the southern United States based on restriction fragment length polymorphism and sequence analysis. J. Clin. Microbiol 39, 2500–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Binder K, Dzaferovic E, Hizo-Teufel C, Sing A, Wildner M, Fingerle V, Jolley KA, 2015. PubMLST.org—the new home for the Borrelia MLSA database. Ticks Tick Borne Dis. 6, 869–871. [DOI] [PubMed] [Google Scholar]
- Margos G, Fedorova N, Kleinjan JE, Hartberger C, Schwan TG, Sing A, Fingerle V, 2017. Borrelia lanei sp. nov. extends the diversity of Borrelia species in California. Int. J. Syst. Evol. Microbiol 67, 3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, Vollmer SA, Cornet M, Piesman J, Donaghy M, Bormane A, Hurn MA, Feil EJ, Fish D, Casjens S, Wormser GP, Schwartz I, Kurtenbach K, 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. U.S.A 105, 8730–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Hojgaard A, Lane RS, Cornet M, Fingerle V, Rudenko N, Ogden N, Aanensen DM, Fish D, Piesman J, 2010. Multilocus sequence analysis of Borrelia bissettii strains from North America reveals a new Borrelia species, Borrelia kurtenbachii. Ticks Tick Borne Dis. 1, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G, Lane RS, Fedorova N, Koloczek J, Piesman J, Hojgaard A, Sing A, Fingerle V, 2016. Borrelia bissettiae sp. nov. and Borrelia californiensis sp. nov. prevail in diverse enzootic transmission cycles. Int. J. Syst. Evol. Microbiol 66 (3), 1447–1452. http://dx.doi.Org/10.1099/ijsem.0.000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, 2015. Epidemiology of Lyme disease. Inf. Dis. Clin. N. Am 29, 187–210. [DOI] [PubMed] [Google Scholar]
- Merten HA, Durden LA, 2000. A state-by-state survey of ticks recorded from humans in the United States. J. Vector Ecol 25, 102–113. [PubMed] [Google Scholar]
- Molloy PJ, Telford SR 3rd, Chowdri HR, Lepore TJ, Gugliotta JL, Weeks KE, Hewins ME, Goethert HK, Berardi VP, 2015. Borrelia miyamotoi disease in the Northeastern United States: a case series. Ann. Int. Med 163, 91–98. [DOI] [PubMed] [Google Scholar]
- Morshed MG, Lee MK, Man S, Fernando K, Wong Q, Hojgaard A, Tang P, Mak S, Henry B, Patrick DM, 2015. Surveillance for Borrelia burgdorferi in Ixodes ticks and small rodents in British Columbia. Vector-Borne Zoonotic Dis. 15, 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DS, Lee X, Larson SR, Johnson DK, Loo T, Paskewitz SM, 2017. Prevalence and distribution of human and tick infections with the Ehrlichia muris-like agent and Anaplasma phagocytophilum in Wisconsin, 2009–2015. Vector-Borne Zoon. Dis 17, 229–236. [DOI] [PubMed] [Google Scholar]
- Nelder MP, Russell CB, Sheehan NJ, Sander B, Moore S, Li Y, Johnson S, Patel SN, Sider D, 2016. Human pathogens associated with the blacklegged tick Ixodes scapularis: a systematic review. Parasites Vectors 9 10.1186/s13071-016-1529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P, Diatta G, Socolovschi C, Mediannikov O, Tall A, Bassene H, Trape JF, Raoult D, 2011. Tick-borne relapsing fever borreliosis, rural Senegal. Emerg. Infect. Dis 17, 883–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson J, Schrumpf ME, Raffel SJ, Porcella SF, Guyard C, Lawrence K, Gherardini FC, Schwan TG, 2007. Purine salvage pathways among Borrelia species. Infect. Immun 75, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic D, Garnier M, Baranton G, 2007. Multilocus sequence analysis of atypical Borrelia burgdorferi sensu lato isolates–description of Borrelia californiensis sp. nov., and genomospecies 1 and 2. Int. J. Med. Microbiol 297, 263–271. [DOI] [PubMed] [Google Scholar]
- Postic D, Ras NM, Lane RS, Hendson M, Baranton G, 1998. Expanded diversity among Californian Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127). J. Clin. Microbiol 36, 3497–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt BS, Allerdice MEJ, Sloan LM, Paddock CD, Munderloh UG, Rikihisa Y, Tajima T, Paskewitz SM, Neitzel DF, Hoang Johnson DK, Schiffman E, Davis JP, Goldsmith CS, Nelson CM, Karpathy SE, 2017. Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int. J. Syst. Evol. Microbiol 67 (7), 2121–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt BS, Mead PS, Johnson DK, Neitzel DF, Respicio-Kingry LB, Davis JP, Schiffman E, Sloan LM, Schriefer ME, Replogle AJ, Paskewitz SM, Ray JA, Bjork J, Steward CR, Deedon A, Lee X, Kingry LC, Miller TK, Feist MA, Theel ES, Patel R, Irish CL, Petersen JM, 2016a. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect. Dis 16, 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt BS, Respicio-Kingry LB, Sloan LM, Schriefer ME, Replogle AJ, Bjork J, Liu G, Kingry LC, Mead PS, Neitzel DF, Schiffman E, Hoang Johnson DK, Davis JP, Paskewitz SM, Boxrud D, Deedon A, Lee X, Miller TK, Feist MA, Steward CR, Theel ES, Patel R, Irish CL, Petersen JM, 2016b. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper midwestern United States. Int. J. Syst. Evol. Microbiol 10.1099/ijsem.0.001445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt BS, Sloan LM, Johnson DK, Munderloh UG, Paskewitz SM, McElroy KM, McFadden JD, Binnicker MJ, Neitzel DF, Liu G, Nicholson WL, Nelson CM, Franson JJ, Martin SA, Cunningham SA, Steward CR, Bogumill K, Bjorgaard ME, Davis JP, McQuiston JH, Warshauer DM, Wilhelm MP, Patel R, Trivedi VA, Eremeeva ME, 2011. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N. Engl. J. Med 365, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SJ, Neitzel DF, Moen RA, Craft ME, Hamilton KE, Johnson LB, Mulla DJ, Munderloh UG, Redig PT, Smith KE, Turner CL, Umber JK, Pelican KM, 2015. Disease risk in a dynamic environment: the spread of tick-borne pathogens in Minnesota, USA. Ecohealth 12, 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Lin T, Gao L, Grubhoffer L, Oliver JH Jr., 2009. Delineation of a new species of the Borrelia burgdorferi sensu lato complex, Borrelia americana sp. nov. J. Clin. Microbiol 47, 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotthoefer AM, Frost HM, 2015. Ecology and epidemiology of Lyme borreliosis. Clin. Lab. Med 35, 723–743. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Battisti JM, Porcella SF, Raffel SJ, Schrumpf ME, Fischer ER, Carroll JA, Stewart PE, Rosa P, Somerville GA, 2003. Glycerol-3-phosphate acquisition in spirochetes: distribution and biological activity of glycerophosphodiester phosphodiesterase (glpQ) among Borrelia species. J. Bacteriol 185, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AM, Hinckley AF, Mead PS, Hook SA, Kugeler KJ, 2017. Surveillance for Lyme disease – United States, 2008–2015. MMWR Surveill. Summ 66, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles GA, Papero M, Beati L, Fish D, 2001. A relapsing fever group spirochete transmitted by Ixodes scapularis ticks. Vector-Borne Zoonotic Dis. 1, 21–34. [DOI] [PubMed] [Google Scholar]
- Souza SS, Bishop HS, Sprinkle P, Qvarnstrom Y, 2016. Comparison of Babesia microti real-time polymerase chain reaction assays for confirmatory diagnosis of babesiosis. Am. J. Trop. Med. Hyg 95, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman A, 1976. Human babesiosis on Nantucket Island: transmission by nymphal Ixodes ticks. Am. J. Trop. Med. Hyg 25, 784–787. [DOI] [PubMed] [Google Scholar]
- Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE, 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med 308, 733–740. [DOI] [PubMed] [Google Scholar]
- Stromdahl E, Hamer S, Jenkins S, Sloan L, Williamson P, Foster E, Nadolny R, Elkins C, Vince M, Pritt B, 2014. Comparison of phenology and pathogen prevalence, including infection with the Ehrlichia muris-like (EML) agent, of Ixodes scapularis removed from soldiers in the midwestern and the northeastern United States over a 15 year period (1997–2012). Parasites Vectors 7 10.1186/s13071-014-0553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglas MB, Foley J, 2006. Differences in the transmissibility of two Anaplasma phagocytophilum strains by the North American tick vector species, Ixodes pacificus and Ixodes scapularis (Acari: Ixodidae). Exp. Appl. Acarol 38, 47–58. [DOI] [PubMed] [Google Scholar]
- Tokarz R, Kapoor V, Samuel JE, Bouyer DH, Briese T, Lipkin WI, 2009. Detection of tick-borne pathogens by MassTag polymerase chain reaction. Vector-Borne Zoonotic Dis. 9, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Tagliafierro T, Cucura DM, Rochlin I, Sameroff S, Lipkin WI, 2017. Detection of Anaplasma phagocytophilum, Babesia microti, Borrelia burgdorferi, Borrelia miyamotoi, and Powassan virus in ticks by a multiplex real-time reverse transcription-PCR assay. mSphere 2 (2). 10.1128/mSphere.00151-17. e00151–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann AJ, Gabitzsch ES, Schulze TL, Zeidner NS, Piesman J, 2005. Three multiplex assays for detection of Borrelia burgdorferi sensu lato and Borrelia miyamotoi sensu lato in field-collected Ixodes nymphs in North America. J. Med. Entomol 42, 1057–1062. [DOI] [PubMed] [Google Scholar]
- Vannier E, Krause PJ, 2012. Human babesiosis. N. Engl. J. Med 366, 2397–2407. [DOI] [PubMed] [Google Scholar]
- Wang G, Liveris D, Mukherjee P, Jungnick S, Margos G, Schwartz I, 2014. Molecular typing of Borrelia burgdorferi. Curr. Protoc. Microbiol 34 10.1002/9780471729259.mc12c05s34. 12C.5.1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski D, Gebhardt L, Prusinski MA, Meehan LJ, Halse TA, Musser KA, 2017. Detection of Borrelia miyamotoi and other tick-borne pathogens in human clinical specimens and Ixodes scapularis ticks in New York State, 2012–2015. Ticks Tick Borne Dis. 8, 407–411. [DOI] [PubMed] [Google Scholar]
- Zeidner NS, Schneider BS, Dolan MC, Piesman J, 2001. An analysis of spirochete load, strain, and pathology in a model of tick-transmitted Lyme borreliosis. Vector-Borne Zoonotic Dis. 1, 35–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.