Figure 3. The yeast RBPome identified by TRAPP compared to poly(A) RNA RBPome.
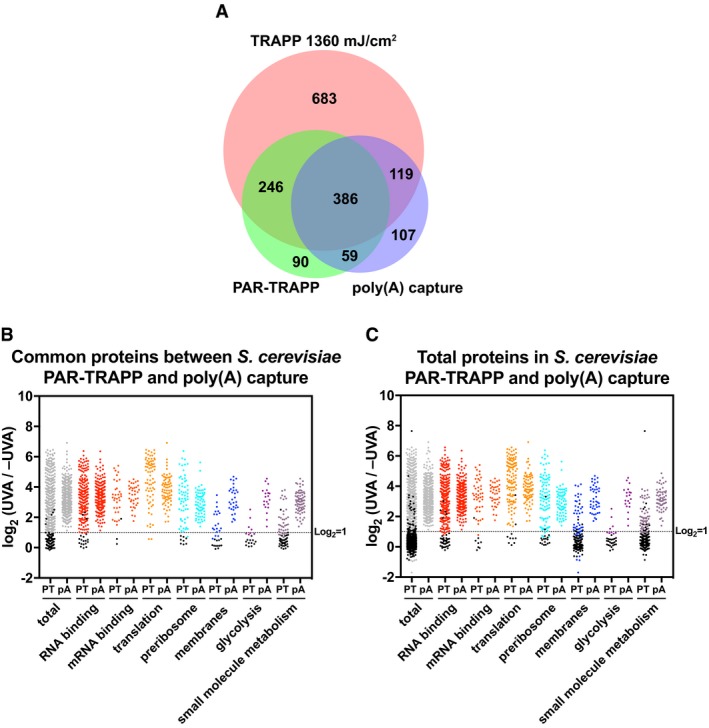
- Venn diagram showing the overlap between proteins identified in PAR‐TRAPP, poly(A) capture and TRAPP.
- Scatter plot of Log2 PAR‐TRAPP SILAC ratios +UVA/−UVA for Saccharomyces cerevisiae proteins and Log2 +UVA/−UVA fold enrichment for poly(A) capture technique. Only proteins identified in both methods as RBPs are shown. Proteins were subdivided based on the indicated GO term categories. Proteins belonging to GO terms “membrane” and “small molecule metabolism” do not contain proteins mapping to GO terms “RNA metabolic process”, “RNA binding”, “ribonucleoprotein complex”. Black dots represent proteins that failed to pass statistical significance cut‐off (P‐value adjusted < 0.05).
- Scatter plot of Log2 SILAC ratios +UVA/−UVA for all S. cerevisiae proteins identified in PAR‐TRAPP plotted together with Log2 +UVA/−UVA fold enrichment for all proteins, reported as RBPs in poly(A) capture technique. Labelling is as in panel (B).
