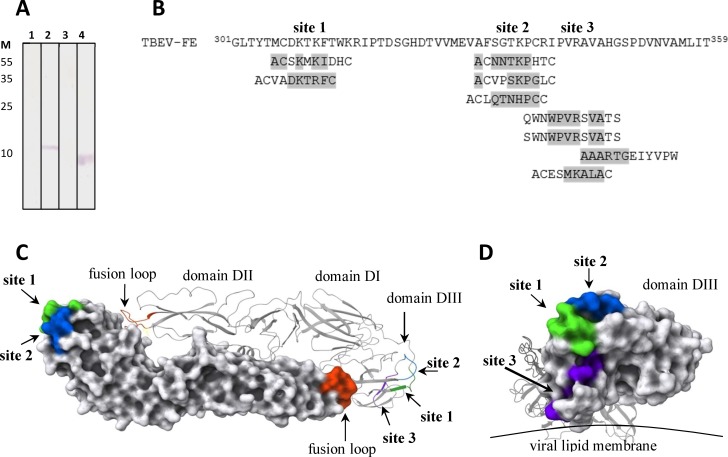
Fig 6. Epitope mapping of chFVN145.
(A) Western blot analysis of lysates of E. coli cells producing rED1+2 (1), rED3_301 (2), rED3delD+ (3) and rED3delA+ (4), which were fractionated by SDS-PAGE (12.5%) and developed with chFVN145. Protein marker masses, in kilodaltons, are shown at the left side of the gel. (B) Results of phage display screening for peptides bound by chFVN145. Alignment of TBEV glycoprotein E homolog fragments and peptides, selected from PhD-12 and PhDC7C library TBEV protein E-like aa was marked in grey. (C) Ribbon and surface representations of TBEV glycoprotein E dimer structure (PDB 1svb) with indicated putative epitope sites. View from the outer surface of virion. (D) Ribbon and surface representations of proposed epitope sites, view from the DIII side of E glycoprotein. Molecular coordinates for the TBEV glycoprotein E structure (PDB 1svb) used in structural analysis were obtained from the Protein Data Bank then visualized using PyMol software (version 1.7.6.0).