Figure 1.
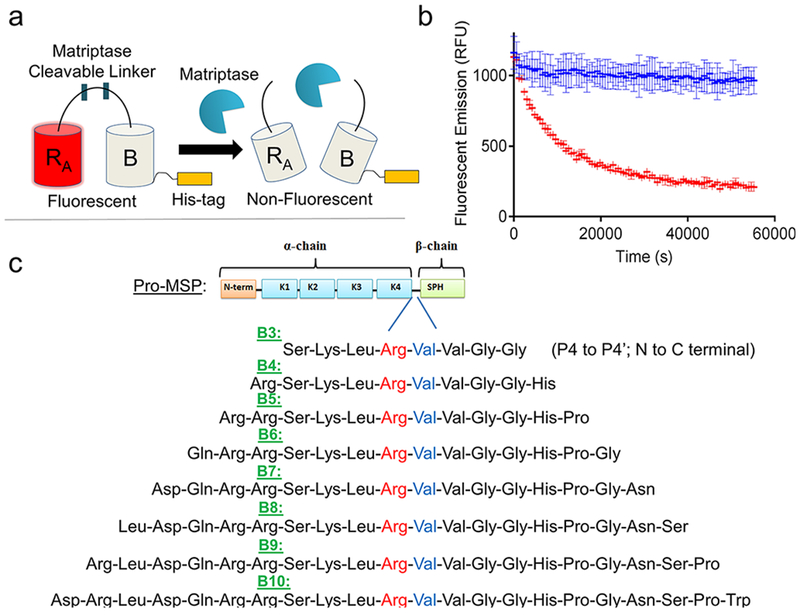
Matriptase biosensor design. (a) ddRFP-based biosensor schematic. Following proteolytic linker cleavage, the fluorescent “A” copy separates from the stabilizing “B” copy, resulting in loss of “A” copy fluorescence. (b) Example time course plot of biosensor mechanism; addition of matriptase cleaves the biosensor and reduces fluorescence over time (red), while absence of matriptase retains fluorescence over time (blue). (c) Matriptase cleavable linker designs and sequence information. Design name is derived from number of amino acids flanking the scissile bond. Scissile bond is highlighted in red (Arg) and blue (Val). Each sequence shown is derived from the natural pro-macrophage stimulating protein (Pro-MSP) sequence. N-term = N-terminus; K1–4 = Kringle domains 1–4; SPH = Serine Protease Homologue.
