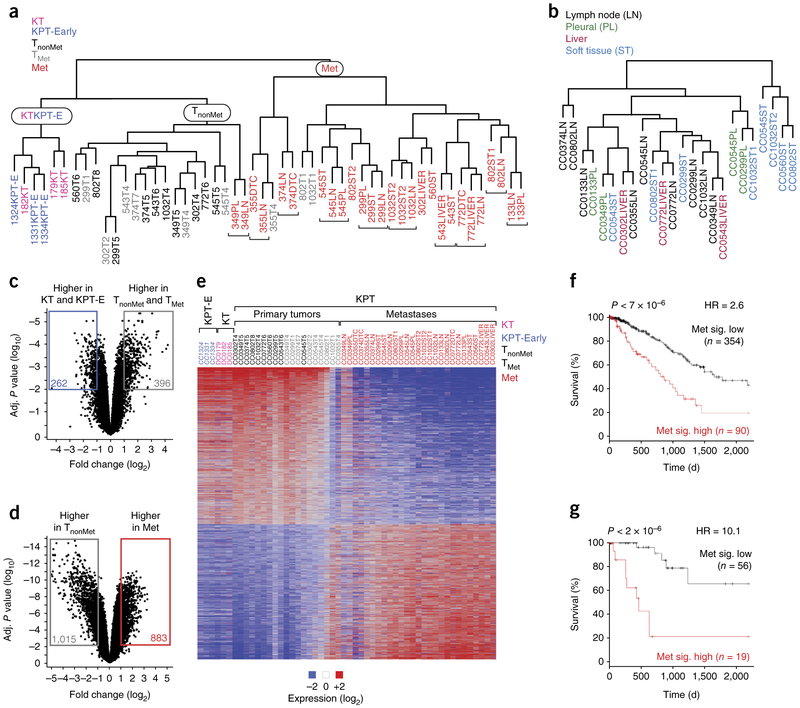
Figure 2.
Lung tumors undergo stepwise changes in gene expression programs during metastatic progression. (a) Clustering analysis of all samples identified three major groups: one with only KT and KPT-E samples, one with all TnonMet samples, and one with almost all (>90%) metastases. Adjacent clonally related metastases are indicated with brackets. (b) Clustering of metastases samples after the gene expression differences that drive clustering of clonally related metastases are removed. (c,d) Robust and conserved changes in gene expression between KT and KPT-E versus primary tumor samples (TnonMet and TMet) (c) and between TnonMet and Met samples (d). The number of genes with >2-fold difference and an adjusted P < 0.01 between groups is indicated. (e) Heat map of genes that are differentially expressed (fold change > 2, adjusted P < 0.01) between TnonMet and Met samples. These samples were not clustered on the basis of their gene expression but rather were arranged by group. For all primary tumors, we dissociated the entire tumor before FACS-based purification of the Tomato+ cancer cells (as opposed to only isolating cells from one part of the tumor). Therefore, the gene expression profile is an average of the entire tumor. Two clonally related metastases 349LN and 349PL were unlike the other metastases. (f,g) Outcome of patients with lung adenocarcinoma after stratification based on a gene expression signature generated by comparing TnonMet to Met samples (Met sig.). Analysis of 444 unselected patients (f) and 75 patients with KRAS-mutant tumors (g) from TCGA. P values, hazard ratios (HR), and number of patients in each group are indicated.