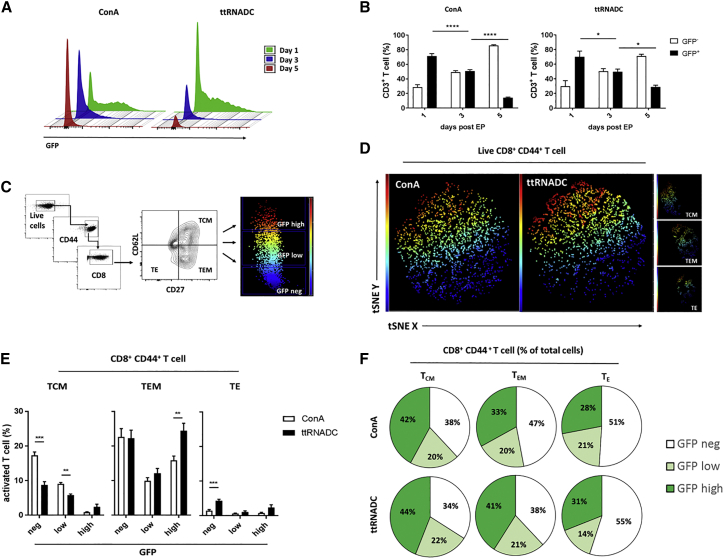
Figure 2.
Phenotypic Analysis of GFP RNA-Modified T Cells
Spleen was harvested from naive or ttRNADC-vaccinated C57BL/6 mice. Single-cell suspension was expanded with ttRNADC or ConA for 5 and 8 days, respectively. Activated T cells were electroporated with 10 μg GFP RNA. Cells were harvested for phenotypic analysis of GFP+ cells by flow cytometry at the indicated time points. (A and B) Representative histogram (A) and percentages of GFP+ CD3+ T cells expanded with ConA and ttRNADC (B). (C) Gating strategy to distinguish GFP expression within TCM (CD62L+, CD27+), TEM (CD62L−, CD27+), and TE (CD62L−, CD27−) compartments under the gate of CD8+ CD44+ T cells. (D and E) Representative t-SNE analysis (D) and percentages of TCM, TEM, and TE (E) negative for GFP or expressing low or high GFP, 24 hr after electroporation. (F) Percentages of CD8+ CD44+ T cell TCM, TEM, and TE negative for GFP or expressing low or high GFP of total cells, 24 hr after electroporation. TCM, central memory T cell; TEM, effector memory T cell; TE, effector T cell; t-SNE, t-distributed stochastic neighbor embedding; ttRNADC, total tumor RNA-pulsed dendritic cell (*p < 0.05; ***p < 0.001; ****p < 0.0001, two-way ANOVA; **p < 0.01, unpaired t test). Values indicated are the mean ± SEM.