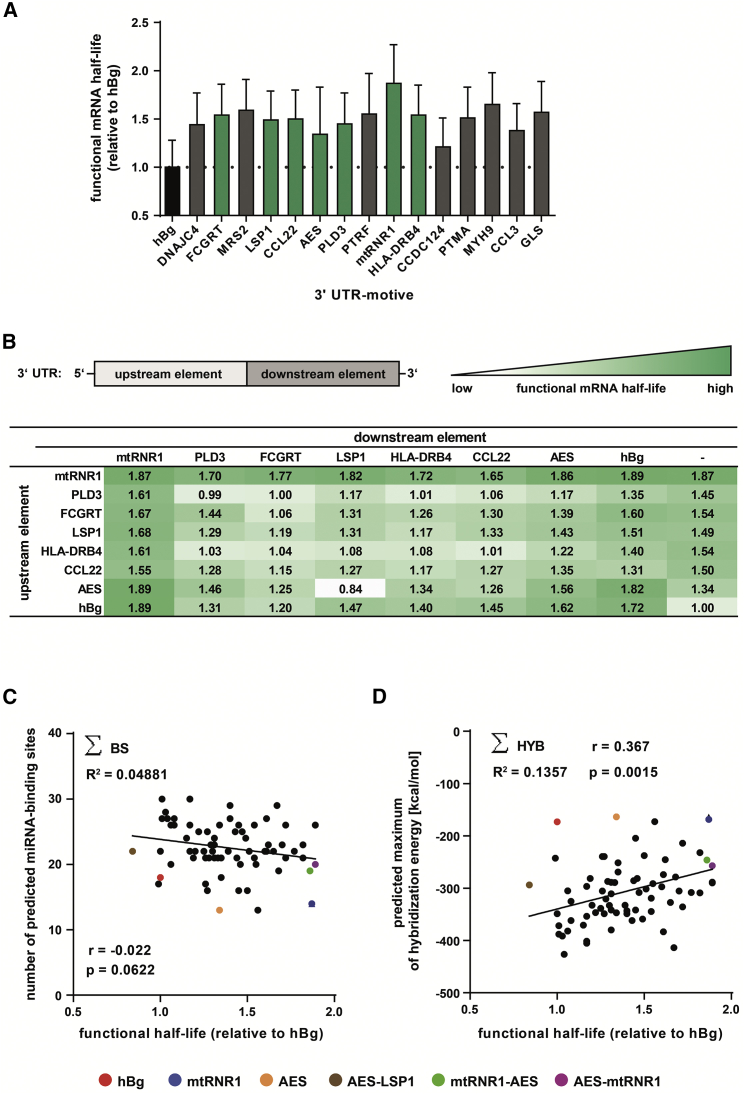
Figure 2.
Identification of 3′ UTR Motives Resulting in Higher Functional Half-Lives of mRNA and Correlation to Predicted miRNA-Binding Sites
(A and B) Sequences of interest were cloned as single (A) or as double (B) 3′ UTR of a destabilized luciferase reporter gene (Luc2CP). Equal amounts of respective mRNAs were electroporated into hDCs, luciferase activity was measured over 72 hr, and functional mRNA half-lives were calculated and compared to the standard hBg 3′ UTR. A summary of 11 experiments is shown. (C and D) Correlation of functional half-lives of mRNAs tagged with single or double 3′ UTR motives, respectively, with the sum of predicted miRNA-binding sites located on the indicated motive (∑BS) (C) and predicted maximum hybridization energy resulting from miRNA-binding sites located on the indicated motive (∑HYB) (D). Putative binding sites for miRNAs and their respective maximum hybridization energies in the sequences of interest were identified via IntaRNA using a miRNA set specific for hDCs. **p < 0.01; n.s., p > 0.05; r, Pearson correlation coefficient.