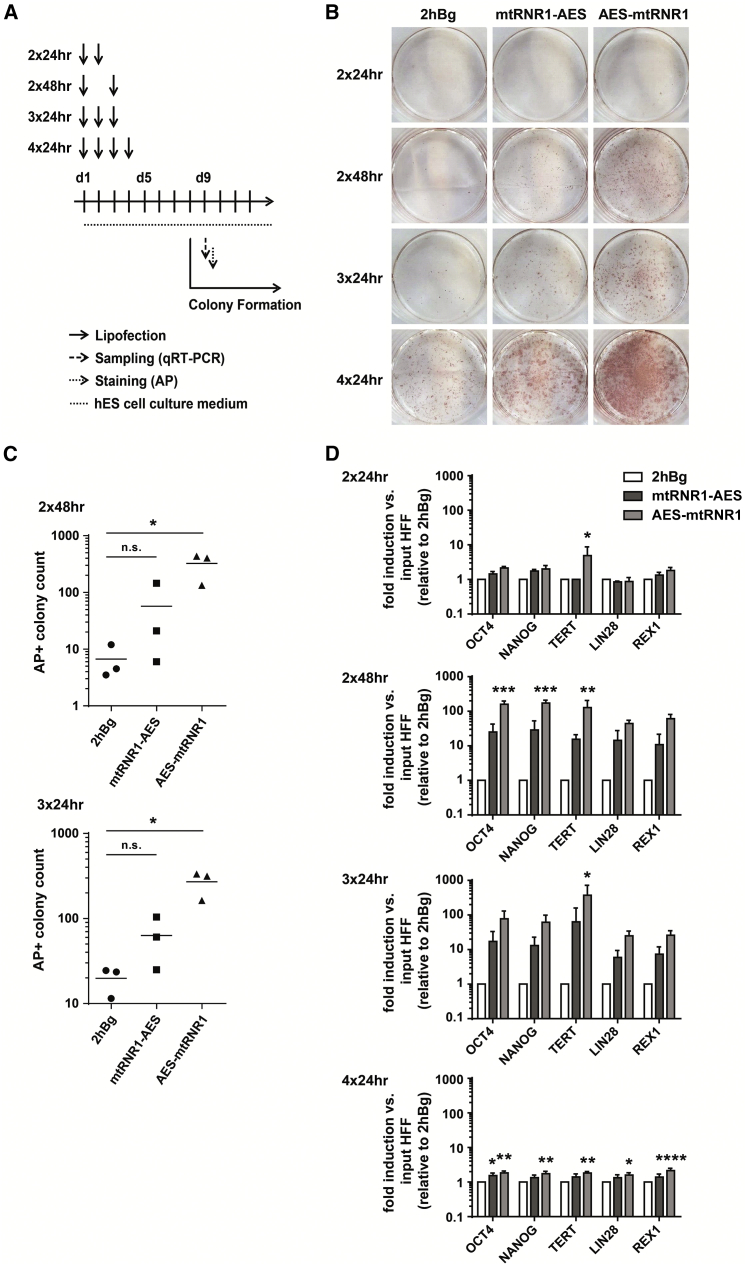
Figure 5.
Improved Reprogramming with mRNAs Tagged with mtRNR1 and AES 3′ UTR Motives
(A) Experimental setup for the reprogramming of human foreskin fibroblasts (HFFs) using synthetic mRNA coding for reprogramming transcription factors, immune evasion proteins, and pluripotency-promoting miRNAs.19, 20 HFFs were lipofected two (2× 24 hr, 2× 48 hr), three (3× 24 hr), or four (4× 24 hr) times. Analysis of colonies was performed on day 9. (B) Representative alkaline phosphatase (AP) staining of HFF-derived iPSCs. (C) Day 9 colony count of AP-positive iPSC colonies comparing two (day 1, day 3) versus three (day 1, day 2, day 3) cycles of lipofection. Colony counts with four lipofections could not be reliably analyzed due to overgrowth and are not shown. One-way ANOVA, Dunnett’s post-test; *p < 0.05; n.s., not significant (three independently performed experiments). (D) Expression of endogenous hESC marker in iPSC colonies was analyzed by qRT-PCR on day 9. Relative expression changes of each transcript in the probe of interest as compared to the day 1 starting control (input HFF) were quantified using the ΔΔCT method and normalized to the housekeeping gene HPRT. The fold inductions obtained by this calculation relative to the corresponding 2hBg benchmark are displayed as mean ± SEM. Two-way ANOVA, Tukey’s post-test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, relative to 2hBg (three independently performed experiments).