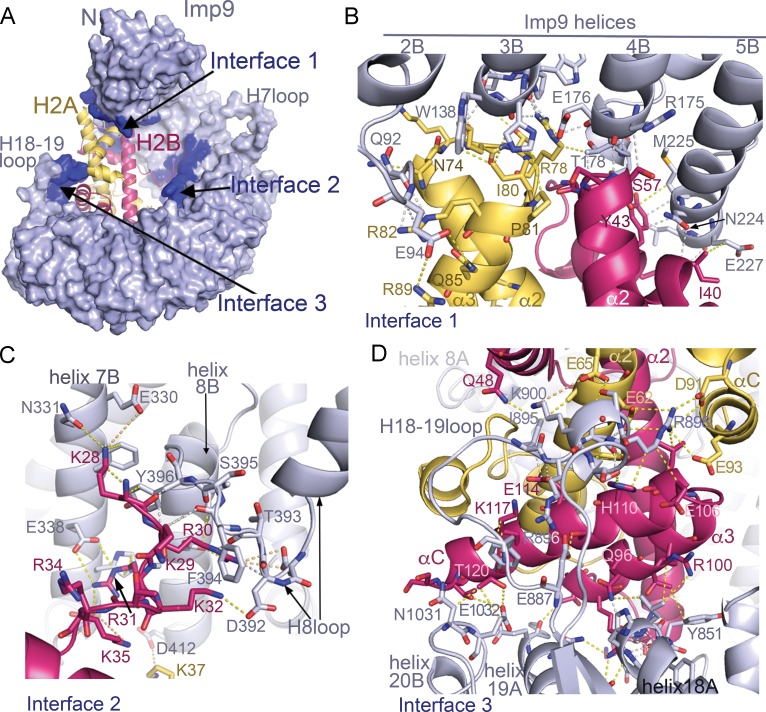
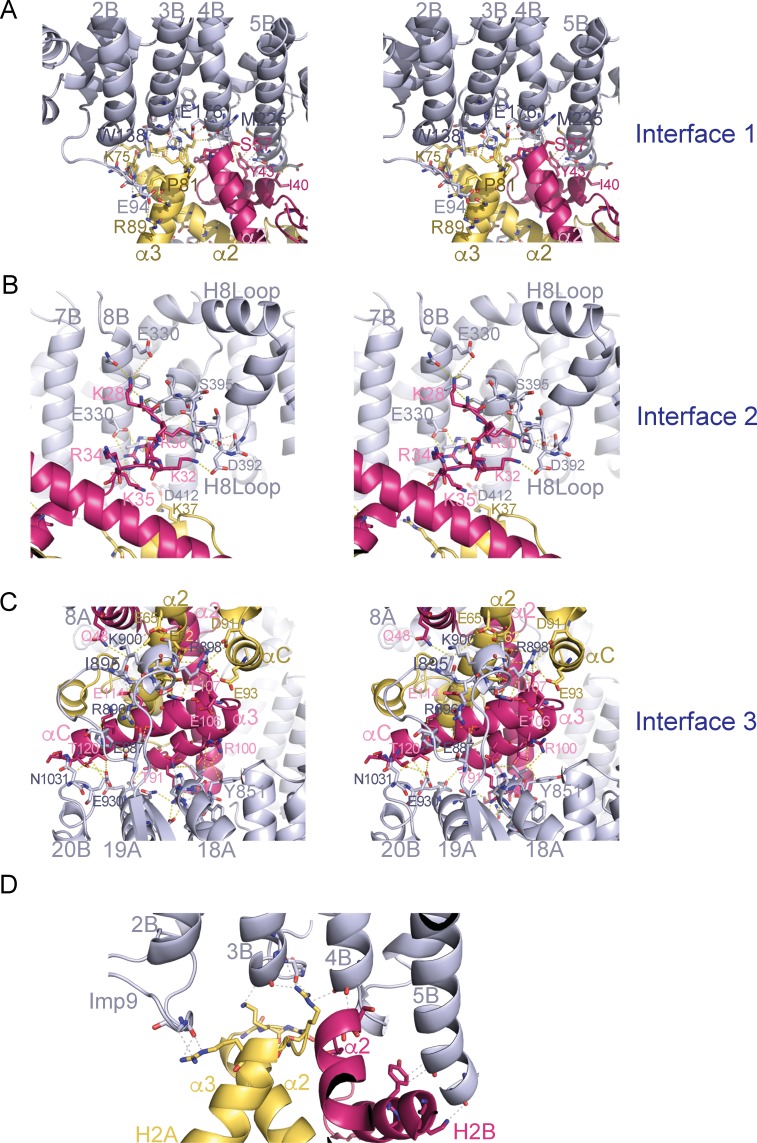
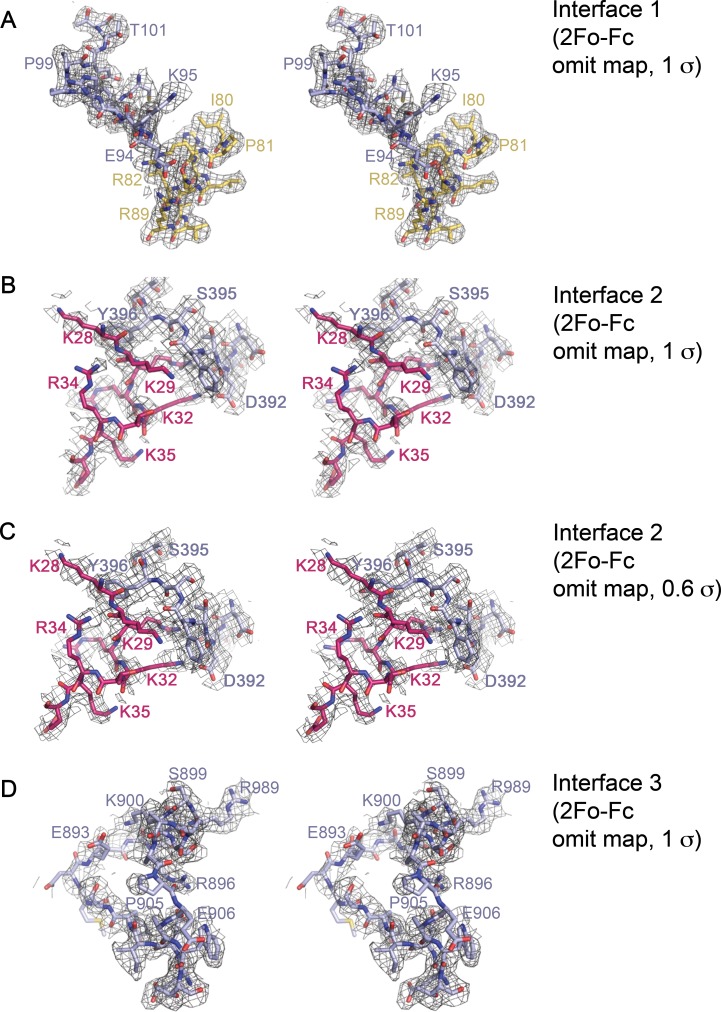
Figure 2. Imp9 •H2A-H2B binding interfaces.
(A) The Imp9•H2A-H2B structure is oriented as in Figure 1C. The histones H2A (yellow)-H2B (red) are drawn as cartoons. Imp9 (blue) is represented as surface showing three distinct H2A-H2B binding interfaces (dark blue). (B–D). Details of Interface 1 (B), Interface 2 (C) and Interface 3 (D). Intermolecular contacts are shown as dashed lines.