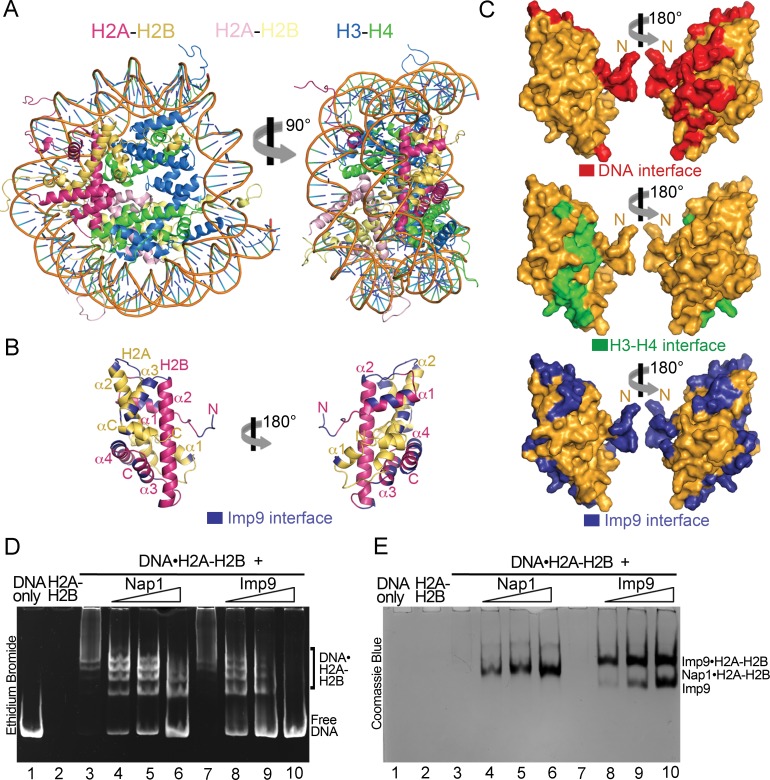
Figure 3. Imp9 has structural and biochemical characteristics of a histone chaperone.
(A) Structure of the nucleosome (1AOI): the orientation on the right shows one of the H2A-H2B dimers (in red and yellow) in the same orientation as H2A-H2B shown in the right panel of B. (B) Imp9-bound H2A-H2B (Imp9 not shown) with its Imp9 interface in dark blue. Orientation of H2A-H2B on the left is the same as in Figures 1C and 2A. (C) Surface representations of the H2A-H2B dimer surface (same orientation as in B) showing nucleosomal DNA (red), nucleosomal H3-H4 (green) and Imp9 (blue) binding interfaces. (D-E) Gel-shift assays to probe chaperone activity of Imp9. Increasing concentrations of Imp9 or Nap1 (0.5, 1.0 and 1.5 molar equivalents of H2A-H2B) were added to pre-formed DNA•H2A-H2B complexes, and the mixtures separated on a native gel stained with ethidium bromide to visualize DNA (D) and with Coomassie Blue to visualize protein (E). The two images of the same gel are horizontally aligned. The histone chaperone Nap1 binds H2A-H2B (E, lanes 4–6) leading to the release of free DNA (D, lanes 4–6). Imp9 also releases free DNA (D, lanes 8–10) as it binds H2A-H2B (E, lanes 8–10).